Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1568-S [Download] |
| Generation | M2-SOS |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1568-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 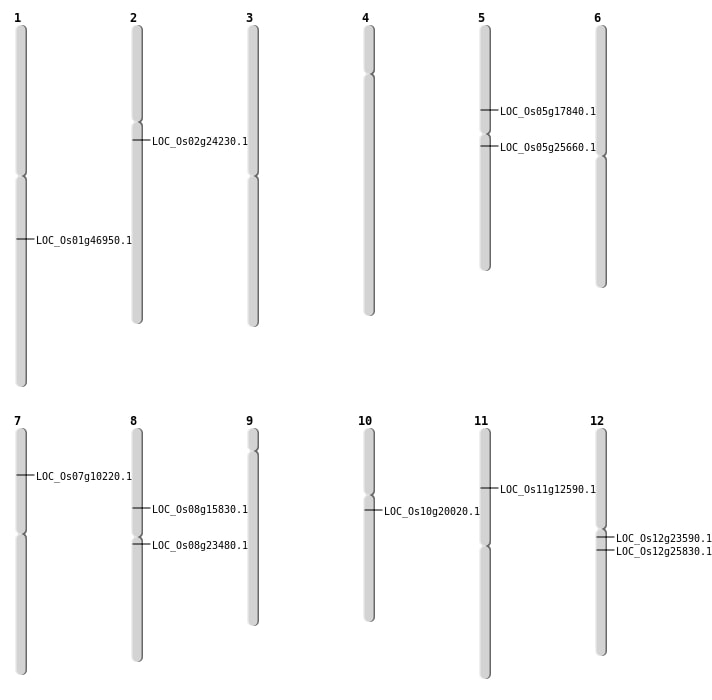
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14149775 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 26792150 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g46950 |
| SBS | Chr1 | 32335278 | G-A | HET | INTRON | |
| SBS | Chr1 | 36423308 | C-T | Homo | INTERGENIC | |
| SBS | Chr1 | 5364970 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 9084020 | A-C | Homo | INTERGENIC | |
| SBS | Chr10 | 10043080 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g20020 |
| SBS | Chr10 | 1820324 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 2066447 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 2484291 | G-A | HET | INTRON | |
| SBS | Chr11 | 17833607 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 18949700 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 14980045 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g25830 |
| SBS | Chr2 | 11041086 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 12223511 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 14042927 | T-A | HET | STOP_GAINED | LOC_Os02g24230 |
| SBS | Chr2 | 33766950 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 9716265 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 10265833 | T-A | HET | INTRON | |
| SBS | Chr3 | 1858419 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 22989017 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 25747445 | A-G | Homo | INTRON | |
| SBS | Chr3 | 9475940 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 10263761 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g17840 |
| SBS | Chr6 | 4532809 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 2356905 | T-C | HET | INTERGENIC | |
| SBS | Chr7 | 5491073 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g10220 |
| SBS | Chr7 | 8768911 | A-T | HET | INTRON | |
| SBS | Chr8 | 15985283 | C-T | HET | INTRON | |
| SBS | Chr8 | 16141389 | C-T | HET | INTRON | |
| SBS | Chr8 | 16142178 | G-A | Homo | SPLICE_SITE_REGION | |
| SBS | Chr8 | 9614618 | C-T | HET | STOP_GAINED | LOC_Os08g15830 |
| SBS | Chr9 | 4239018 | G-A | HET | INTRON | |
| SBS | Chr9 | 5731802 | G-T | Homo | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 4118059 | 4118060 | 1 | |
| Deletion | Chr10 | 4586821 | 4586822 | 1 | |
| Deletion | Chr10 | 4913981 | 4913982 | 1 | |
| Deletion | Chr9 | 6658819 | 6658820 | 1 | |
| Deletion | Chr6 | 7220499 | 7220500 | 1 | |
| Deletion | Chr12 | 7329412 | 7329413 | 1 | |
| Deletion | Chr4 | 7978559 | 7978560 | 1 | |
| Deletion | Chr11 | 8194440 | 8194441 | 1 | |
| Deletion | Chr10 | 9876252 | 9876254 | 2 | |
| Deletion | Chr8 | 11377104 | 11377109 | 5 | |
| Deletion | Chr12 | 12051331 | 12051332 | 1 | |
| Deletion | Chr11 | 13011947 | 13011955 | 8 | |
| Deletion | Chr8 | 14218219 | 14218236 | 17 | LOC_Os08g23480 |
| Deletion | Chr8 | 14891261 | 14891262 | 1 | |
| Deletion | Chr4 | 17471541 | 17471550 | 9 | |
| Deletion | Chr7 | 19774622 | 19774623 | 1 | |
| Deletion | Chr12 | 21009930 | 21009931 | 1 | |
| Deletion | Chr1 | 23103354 | 23103364 | 10 | |
| Deletion | Chr3 | 23513678 | 23513685 | 7 | |
| Deletion | Chr11 | 23963378 | 23963390 | 12 | |
| Deletion | Chr3 | 26820029 | 26820036 | 7 | |
| Deletion | Chr5 | 28229700 | 28229701 | 1 | |
| Deletion | Chr2 | 29687523 | 29687524 | 1 | |
| Deletion | Chr1 | 32543954 | 32543969 | 15 | |
| Deletion | Chr3 | 33875862 | 33875867 | 5 | |
| Deletion | Chr1 | 34523434 | 34523435 | 1 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 27933585 | 27933586 | 2 | |
| Insertion | Chr1 | 30276415 | 30276416 | 2 | |
| Insertion | Chr1 | 4634006 | 4634030 | 25 | |
| Insertion | Chr1 | 7284691 | 7284698 | 8 | |
| Insertion | Chr11 | 7072743 | 7072743 | 1 | LOC_Os11g12590 |
| Insertion | Chr12 | 5665594 | 5665594 | 1 | |
| Insertion | Chr4 | 8607096 | 8607136 | 41 | |
| Insertion | Chr5 | 17503641 | 17503682 | 42 | |
| Insertion | Chr6 | 25885779 | 25885786 | 8 | |
| Insertion | Chr8 | 16866911 | 16866912 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 8487738 | 8741108 | |
| Inversion | Chr7 | 8487751 | 8602892 | |
| Inversion | Chr12 | 8542550 | 13372020 | LOC_Os12g23590 |
| Inversion | Chr5 | 14921597 | 18954040 | LOC_Os05g25660 |
| Inversion | Chr5 | 14933263 | 18954034 | |
| Inversion | Chr7 | 18675147 | 19334772 |
No Translocation
