Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1571-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1571-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 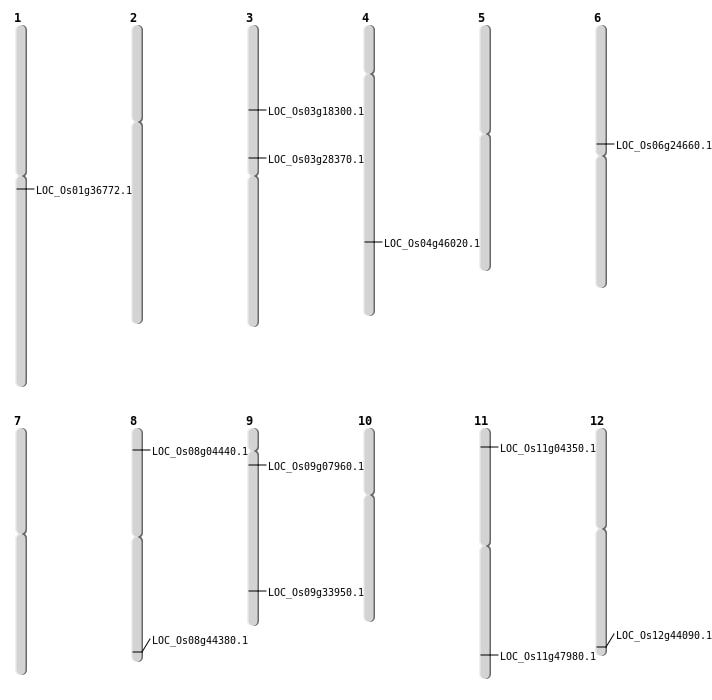
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1126813 | G-T | HET | INTERGENIC | |
| SBS | Chr1 | 13916769 | G-C | Homo | INTERGENIC | |
| SBS | Chr1 | 14487801 | A-G | Homo | INTERGENIC | |
| SBS | Chr1 | 1831793 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 3008245 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 31955079 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 11287859 | A-G | Homo | INTERGENIC | |
| SBS | Chr10 | 15189448 | T-G | HET | INTERGENIC | |
| SBS | Chr10 | 15189449 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 16668669 | C-T | HET | INTRON | |
| SBS | Chr10 | 18395524 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 26409520 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 17522702 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 17974736 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 25214045 | A-G | HET | INTRON | |
| SBS | Chr3 | 10005734 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14189801 | G-A | HET | INTRON | |
| SBS | Chr3 | 18495590 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 1906691 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 2281893 | T-C | Homo | INTERGENIC | |
| SBS | Chr3 | 27481391 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 27481392 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 6877268 | T-A | HET | INTERGENIC | |
| SBS | Chr3 | 9997044 | A-G | HET | INTRON | |
| SBS | Chr4 | 20434368 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 20434369 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 20434373 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 2245520 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 30170413 | C-T | HET | INTRON | |
| SBS | Chr5 | 10457148 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 12922910 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 325170 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 4086803 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07960 |
Deletions: 42
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 238229 | 238231 | 2 | |
| Deletion | Chr11 | 1363303 | 1363324 | 21 | |
| Deletion | Chr11 | 1809034 | 1809035 | 1 | LOC_Os11g04350 |
| Deletion | Chr8 | 2175032 | 2175039 | 7 | LOC_Os08g04440 |
| Deletion | Chr12 | 2808835 | 2808869 | 34 | |
| Deletion | Chr6 | 7348372 | 7348375 | 3 | |
| Deletion | Chr11 | 7349882 | 7349883 | 1 | |
| Deletion | Chr12 | 7495363 | 7495371 | 8 | |
| Deletion | Chr4 | 8190398 | 8190399 | 1 | |
| Deletion | Chr10 | 8231859 | 8231860 | 1 | |
| Deletion | Chr1 | 8321270 | 8321281 | 11 | |
| Deletion | Chr2 | 8632496 | 8632499 | 3 | |
| Deletion | Chr10 | 9161227 | 9161229 | 2 | |
| Deletion | Chr3 | 9989469 | 9989470 | 1 | |
| Deletion | Chr2 | 10221075 | 10221081 | 6 | |
| Deletion | Chr7 | 10665390 | 10665391 | 1 | |
| Deletion | Chr7 | 12223446 | 12235672 | 12226 | |
| Deletion | Chr3 | 13018985 | 13018986 | 1 | |
| Deletion | Chr3 | 13475318 | 13475321 | 3 | |
| Deletion | Chr9 | 14827526 | 14827528 | 2 | |
| Deletion | Chr1 | 15661808 | 15661819 | 11 | |
| Deletion | Chr5 | 16224113 | 16224114 | 1 | |
| Deletion | Chr3 | 16351704 | 16351705 | 1 | LOC_Os03g28370 |
| Deletion | Chr12 | 16727760 | 16727761 | 1 | |
| Deletion | Chr5 | 18612328 | 18612334 | 6 | |
| Deletion | Chr3 | 18631767 | 18631799 | 32 | |
| Deletion | Chr12 | 18751387 | 18751405 | 18 | |
| Deletion | Chr8 | 19727688 | 19727689 | 1 | |
| Deletion | Chr3 | 20002203 | 20002227 | 24 | |
| Deletion | Chr9 | 20040241 | 20040253 | 12 | LOC_Os09g33950 |
| Deletion | Chr1 | 20429513 | 20429531 | 18 | LOC_Os01g36772 |
| Deletion | Chr5 | 21031185 | 21031256 | 71 | |
| Deletion | Chr6 | 21047017 | 21047020 | 3 | |
| Deletion | Chr1 | 21562417 | 21562418 | 1 | |
| Deletion | Chr8 | 24156679 | 24156680 | 1 | |
| Deletion | Chr3 | 25261134 | 25261135 | 1 | |
| Deletion | Chr4 | 27261513 | 27261518 | 5 | LOC_Os04g46020 |
| Deletion | Chr8 | 27930768 | 27930773 | 5 | LOC_Os08g44380 |
| Deletion | Chr1 | 31932468 | 31932469 | 1 | |
| Deletion | Chr1 | 31955030 | 31955033 | 3 | |
| Deletion | Chr4 | 34591195 | 34591202 | 7 | |
| Deletion | Chr1 | 37689189 | 37689198 | 9 |
Insertions: 12
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 10255782 | 30025390 | LOC_Os03g18300 |
| Inversion | Chr6 | 14486314 | 14970366 | LOC_Os06g24660 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 27344056 | Chr11 | 28939329 | 2 |
