Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1586-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1586-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 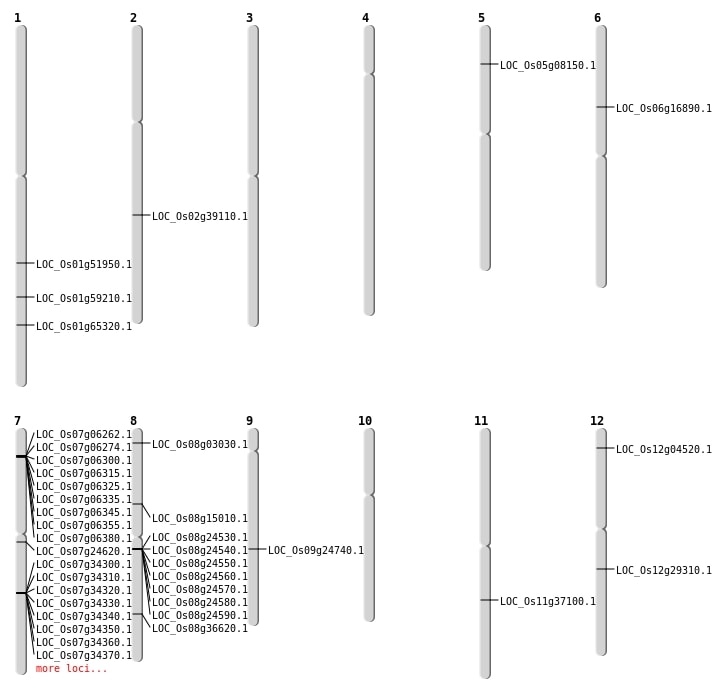
|
| Variant Information |
|---|
Single base substitutions: 29
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 34205579 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g59210 |
| SBS | Chr10 | 294026 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 23604662 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 393436 | T-A | HET | INTRON | |
| SBS | Chr11 | 488269 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 17371484 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g29310 |
| SBS | Chr12 | 1926211 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g04520 |
| SBS | Chr12 | 25385639 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 18426197 | T-C | Homo | INTERGENIC | |
| SBS | Chr2 | 23621980 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g39110 |
| SBS | Chr2 | 23621984 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 27636276 | T-C | Homo | INTERGENIC | |
| SBS | Chr3 | 34638863 | C-G | Homo | INTERGENIC | |
| SBS | Chr4 | 12849927 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 13132910 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 14579533 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 21608677 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 27797511 | A-T | Homo | INTRON | |
| SBS | Chr6 | 4018278 | C-T | HET | INTRON | |
| SBS | Chr6 | 9790648 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g16890 |
| SBS | Chr7 | 14007907 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g24620 |
| SBS | Chr7 | 506126 | C-G | HET | INTERGENIC | |
| SBS | Chr7 | 7174848 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 13106947 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 1351271 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g03030 |
| SBS | Chr8 | 15315065 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 23106305 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g36620 |
| SBS | Chr8 | 9718387 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 14738867 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g24740 |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 371492 | 371499 | 7 | |
| Deletion | Chr1 | 709445 | 709451 | 6 | |
| Deletion | Chr6 | 2882836 | 2882842 | 6 | |
| Deletion | Chr7 | 3042001 | 3105000 | 63000 | 9 |
| Deletion | Chr7 | 3950282 | 3950283 | 1 | |
| Deletion | Chr8 | 6027700 | 6027706 | 6 | |
| Deletion | Chr12 | 6283737 | 6283738 | 1 | |
| Deletion | Chr12 | 7140688 | 7140693 | 5 | |
| Deletion | Chr7 | 8373950 | 8373954 | 4 | |
| Deletion | Chr12 | 13390241 | 13390242 | 1 | |
| Deletion | Chr3 | 13471240 | 13471247 | 7 | |
| Deletion | Chr2 | 13656529 | 13656531 | 2 | |
| Deletion | Chr1 | 14285900 | 14285905 | 5 | |
| Deletion | Chr8 | 14812001 | 14875000 | 63000 | 7 |
| Deletion | Chr8 | 14816755 | 14816770 | 15 | |
| Deletion | Chr8 | 17267701 | 17267703 | 2 | |
| Deletion | Chr12 | 17569359 | 17569360 | 1 | |
| Deletion | Chr12 | 18435767 | 18435788 | 21 | |
| Deletion | Chr7 | 20542001 | 20607000 | 65000 | 9 |
| Deletion | Chr1 | 21258678 | 21258679 | 1 | |
| Deletion | Chr4 | 21716808 | 21716809 | 1 | |
| Deletion | Chr2 | 27695966 | 27695968 | 2 | |
| Deletion | Chr1 | 29863655 | 29863656 | 1 | LOC_Os01g51950 |
| Deletion | Chr1 | 37894475 | 37894478 | 3 | LOC_Os01g65320 |
Insertions: 11
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 4442139 | Chr3 | 31311796 | LOC_Os05g08150 |
| Translocation | Chr5 | 4442420 | Chr3 | 31311795 | LOC_Os05g08150 |
| Translocation | Chr11 | 21907918 | Chr8 | 9057180 | 2 |
