Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1590-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1590-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 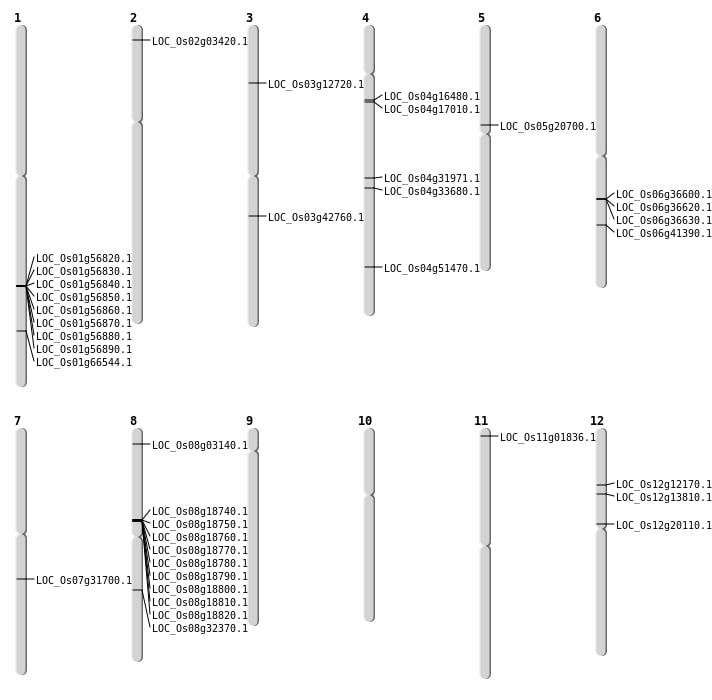
|
| Variant Information |
|---|
Single base substitutions: 17
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 30948005 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 9020077 | G-C | Homo | INTERGENIC | |
| SBS | Chr11 | 20481087 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 20481088 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 11692276 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g20110 |
| SBS | Chr12 | 19313299 | C-T | Homo | INTERGENIC | |
| SBS | Chr12 | 25059169 | T-C | HET | INTRON | |
| SBS | Chr2 | 17594977 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 22315536 | T-G | Homo | INTERGENIC | |
| SBS | Chr3 | 33899586 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr4 | 30401417 | G-C | HET | INTRON | |
| SBS | Chr5 | 18308211 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 16595957 | T-A | Homo | INTRON | |
| SBS | Chr8 | 19263994 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 21750557 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 28107877 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 15379577 | C-T | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 447236 | 447242 | 6 | LOC_Os11g01836 |
| Deletion | Chr2 | 1395414 | 1395415 | 1 | LOC_Os02g03420 |
| Deletion | Chr8 | 1441092 | 1441093 | 1 | LOC_Os08g03140 |
| Deletion | Chr2 | 2446164 | 2446167 | 3 | |
| Deletion | Chr9 | 3062863 | 3062864 | 1 | |
| Deletion | Chr4 | 4101979 | 4102005 | 26 | |
| Deletion | Chr1 | 5642315 | 5642336 | 21 | |
| Deletion | Chr11 | 5721457 | 5721468 | 11 | |
| Deletion | Chr9 | 6759971 | 6759987 | 16 | |
| Deletion | Chr12 | 7807338 | 7807339 | 1 | LOC_Os12g13810 |
| Deletion | Chr3 | 8350160 | 8350178 | 18 | |
| Deletion | Chr4 | 8958498 | 8958502 | 4 | LOC_Os04g16480 |
| Deletion | Chr9 | 10321008 | 10321010 | 2 | |
| Deletion | Chr8 | 11189001 | 11231000 | 42000 | 9 |
| Deletion | Chr5 | 12137245 | 12137253 | 8 | LOC_Os05g20700 |
| Deletion | Chr8 | 13236002 | 13236003 | 1 | |
| Deletion | Chr5 | 13389535 | 13389540 | 5 | LOC_Os05g23430 |
| Deletion | Chr8 | 13695349 | 13695354 | 5 | |
| Deletion | Chr7 | 18821394 | 18821397 | 3 | LOC_Os07g31700 |
| Deletion | Chr4 | 19148980 | 19148994 | 14 | LOC_Os04g31971 |
| Deletion | Chr8 | 20048150 | 20048154 | 4 | LOC_Os08g32370 |
| Deletion | Chr6 | 21503903 | 21522448 | 18545 | 3 |
| Deletion | Chr6 | 21580025 | 21580038 | 13 | |
| Deletion | Chr8 | 22479002 | 22479005 | 3 | |
| Deletion | Chr10 | 23034293 | 23034295 | 2 | |
| Deletion | Chr3 | 23708104 | 23708109 | 5 | |
| Deletion | Chr3 | 23816357 | 23816359 | 2 | LOC_Os03g42760 |
| Deletion | Chr7 | 23945832 | 23945833 | 1 | |
| Deletion | Chr8 | 24342954 | 24342956 | 2 | |
| Deletion | Chr6 | 24816119 | 24816121 | 2 | LOC_Os06g41390 |
| Deletion | Chr2 | 27097510 | 27097514 | 4 | |
| Deletion | Chr7 | 29692569 | 29692571 | 2 | |
| Deletion | Chr6 | 29915734 | 29915735 | 1 | |
| Deletion | Chr4 | 30493983 | 30493984 | 1 | LOC_Os04g51470 |
| Deletion | Chr1 | 32813001 | 32860000 | 47000 | 8 |
| Deletion | Chr1 | 41158600 | 41158605 | 5 | LOC_Os01g71106 |
Insertions: 10
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 20422341 | 20422341 | 1 | |
| Insertion | Chr11 | 22360207 | 22360229 | 23 | |
| Insertion | Chr2 | 24310567 | 24310567 | 1 | |
| Insertion | Chr4 | 20394930 | 20394930 | 1 | LOC_Os04g33680 |
| Insertion | Chr6 | 23287977 | 23287977 | 1 | |
| Insertion | Chr6 | 29432069 | 29432070 | 2 | |
| Insertion | Chr7 | 11342964 | 11343014 | 51 | |
| Insertion | Chr8 | 20412977 | 20412978 | 2 | |
| Insertion | Chr8 | 22800889 | 22800889 | 1 | |
| Insertion | Chr8 | 2626621 | 2626624 | 4 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 1054102 | 9311991 | LOC_Os04g17010 |
| Inversion | Chr4 | 2692695 | 9311988 | LOC_Os04g17010 |
| Inversion | Chr1 | 38168586 | 38644949 | LOC_Os01g66544 |
| Inversion | Chr1 | 38168616 | 38644959 | LOC_Os01g66544 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 6675393 | Chr1 | 32812326 | |
| Translocation | Chr12 | 6679736 | Chr3 | 7847583 | LOC_Os12g12170 |
| Translocation | Chr3 | 6787041 | Chr1 | 14175981 | LOC_Os03g12720 |
