Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1600-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1600-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 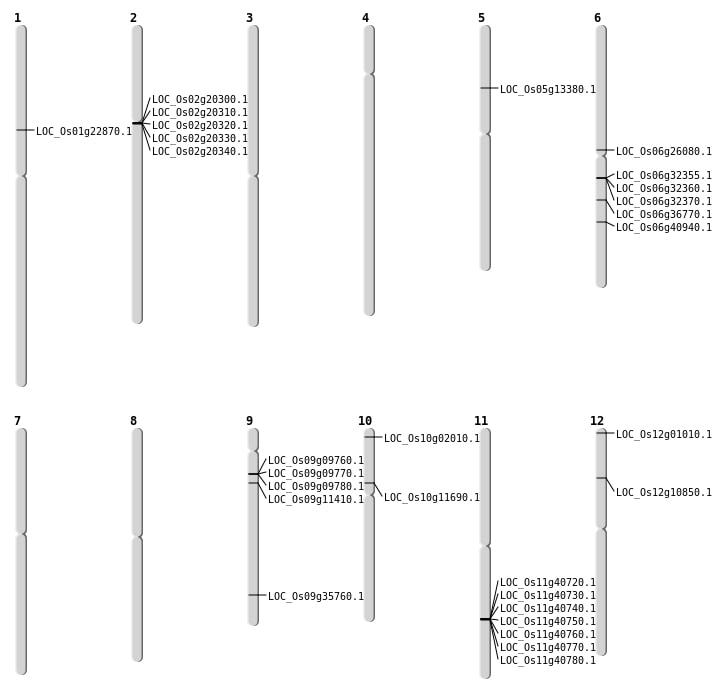
|
| Variant Information |
|---|
Single base substitutions: 24
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 19777518 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29842025 | A-T | HET | INTERGENIC | |
| SBS | Chr1 | 33444629 | G-A | Homo | INTRON | |
| SBS | Chr10 | 20803078 | A-G | HET | INTRON | |
| SBS | Chr10 | 5759614 | T-A | HET | INTERGENIC | |
| SBS | Chr10 | 637239 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g02010 |
| SBS | Chr10 | 6463596 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g11690 |
| SBS | Chr11 | 15445986 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 16893442 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 2480126 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 564367 | T-C | HET | UTR_5_PRIME | |
| SBS | Chr12 | 16750359 | A-T | Homo | INTERGENIC | |
| SBS | Chr2 | 19682271 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 29392176 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 33235833 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 8189832 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 15342943 | A-C | Homo | INTERGENIC | |
| SBS | Chr3 | 15342944 | T-C | Homo | INTERGENIC | |
| SBS | Chr3 | 35866211 | G-A | HET | INTRON | |
| SBS | Chr4 | 11407943 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 5498994 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 8586477 | G-A | HET | INTRON | |
| SBS | Chr6 | 15266175 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g26080 |
| SBS | Chr9 | 16772153 | G-T | HET | INTERGENIC |
Deletions: 41
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 1415531 | 1415535 | 4 | |
| Deletion | Chr2 | 1467368 | 1467372 | 4 | |
| Deletion | Chr3 | 1491340 | 1491344 | 4 | |
| Deletion | Chr11 | 2354353 | 2354354 | 1 | |
| Deletion | Chr2 | 3356298 | 3356300 | 2 | |
| Deletion | Chr3 | 3595127 | 3595129 | 2 | |
| Deletion | Chr10 | 4102247 | 4102261 | 14 | |
| Deletion | Chr9 | 5280352 | 5303631 | 23279 | 3 |
| Deletion | Chr10 | 5367294 | 5367301 | 7 | |
| Deletion | Chr9 | 6354543 | 6354561 | 18 | LOC_Os09g11410 |
| Deletion | Chr10 | 6481136 | 6481137 | 1 | |
| Deletion | Chr5 | 7427225 | 7430847 | 3622 | LOC_Os05g13380 |
| Deletion | Chr5 | 7861073 | 7861082 | 9 | |
| Deletion | Chr4 | 8884411 | 8884412 | 1 | |
| Deletion | Chr10 | 8961248 | 8961263 | 15 | |
| Deletion | Chr2 | 11958284 | 11984963 | 26679 | 5 |
| Deletion | Chr11 | 12315121 | 12315276 | 155 | |
| Deletion | Chr12 | 13980016 | 13980028 | 12 | |
| Deletion | Chr3 | 15234873 | 15234876 | 3 | |
| Deletion | Chr11 | 15298543 | 15298544 | 1 | |
| Deletion | Chr12 | 16305205 | 16305212 | 7 | |
| Deletion | Chr12 | 17185291 | 17185292 | 1 | |
| Deletion | Chr3 | 18547894 | 18547936 | 42 | |
| Deletion | Chr6 | 18830254 | 18839744 | 9490 | 3 |
| Deletion | Chr9 | 19527997 | 19528001 | 4 | |
| Deletion | Chr6 | 20181073 | 20181079 | 6 | |
| Deletion | Chr7 | 20422738 | 20422739 | 1 | |
| Deletion | Chr9 | 20573822 | 20573835 | 13 | LOC_Os09g35760 |
| Deletion | Chr8 | 20772328 | 20772337 | 9 | |
| Deletion | Chr5 | 21547093 | 21547096 | 3 | |
| Deletion | Chr12 | 21855094 | 21855100 | 6 | |
| Deletion | Chr6 | 22472581 | 22472587 | 6 | |
| Deletion | Chr11 | 24355001 | 24396000 | 41000 | 7 |
| Deletion | Chr3 | 24388552 | 24388556 | 4 | |
| Deletion | Chr6 | 24401827 | 24401842 | 15 | LOC_Os06g40940 |
| Deletion | Chr4 | 24451317 | 24451319 | 2 | |
| Deletion | Chr6 | 27225149 | 27225155 | 6 | |
| Deletion | Chr3 | 28147971 | 28147972 | 1 | |
| Deletion | Chr6 | 29909392 | 29909412 | 20 | |
| Deletion | Chr4 | 30520189 | 30520194 | 5 | |
| Deletion | Chr3 | 34944900 | 34944907 | 7 |
Insertions: 9
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 2719 | 2838 | LOC_Os12g01010 |
| Inversion | Chr4 | 11204394 | 11204674 | |
| Inversion | Chr1 | 12849747 | 12849873 | LOC_Os01g22870 |
| Inversion | Chr1 | 19600503 | 22364786 | |
| Inversion | Chr3 | 20164316 | 20434204 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 5851470 | Chr9 | 22371341 | 2 |
| Translocation | Chr4 | 15896406 | Chr1 | 9105547 | |
| Translocation | Chr8 | 20853774 | Chr1 | 17838357 | |
| Translocation | Chr6 | 21657465 | Chr4 | 17923255 | LOC_Os06g36770 |
