Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1606-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1606-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 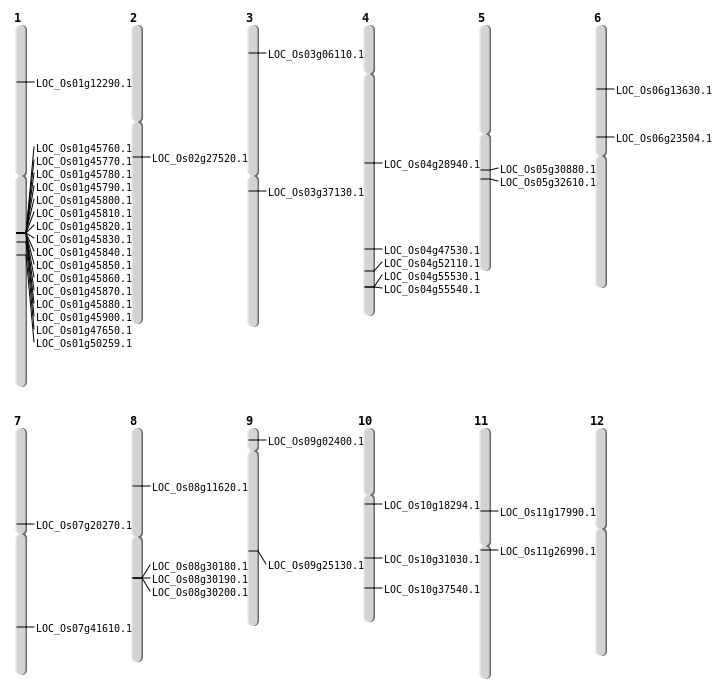
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24912376 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 32570228 | A-G | Homo | INTERGENIC | |
| SBS | Chr10 | 12101135 | G-A | Homo | INTERGENIC | |
| SBS | Chr10 | 13811219 | C-G | HET | INTERGENIC | |
| SBS | Chr10 | 1521065 | G-T | HET | INTERGENIC | |
| SBS | Chr10 | 1521066 | A-T | HET | INTERGENIC | |
| SBS | Chr10 | 20098944 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g37540 |
| SBS | Chr10 | 8600367 | G-C | Homo | INTERGENIC | |
| SBS | Chr11 | 10111159 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17990 |
| SBS | Chr11 | 14601258 | T-G | HET | INTERGENIC | |
| SBS | Chr11 | 15118666 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 18840209 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 22839130 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 8153564 | G-T | HET | INTERGENIC | |
| SBS | Chr2 | 23347680 | G-A | HET | INTRON | |
| SBS | Chr2 | 27604955 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 30499378 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 11531601 | G-A | Homo | INTRON | |
| SBS | Chr3 | 3062859 | G-A | HET | STOP_GAINED | LOC_Os03g06110 |
| SBS | Chr4 | 17156330 | C-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g28940 |
| SBS | Chr4 | 24421969 | T-C | Homo | INTERGENIC | |
| SBS | Chr4 | 2472892 | A-T | Homo | INTERGENIC | |
| SBS | Chr4 | 32960967 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 32960968 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 12323574 | A-G | Homo | INTERGENIC | |
| SBS | Chr5 | 17932767 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g30880 |
| SBS | Chr5 | 8576398 | G-A | Homo | INTRON | |
| SBS | Chr6 | 17552267 | G-T | Homo | INTRON | |
| SBS | Chr6 | 27865070 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 27865071 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 31199284 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14381654 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 29004699 | C-T | HET | INTRON | |
| SBS | Chr7 | 6730941 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 10236866 | A-G | Homo | INTERGENIC | |
| SBS | Chr9 | 1605135 | G-T | Homo | INTERGENIC |
Deletions: 40
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 524553 | 524566 | 13 | |
| Deletion | Chr8 | 2346895 | 2346898 | 3 | |
| Deletion | Chr8 | 2607364 | 2607365 | 1 | |
| Deletion | Chr3 | 2672178 | 2672202 | 24 | |
| Deletion | Chr9 | 2981590 | 2981591 | 1 | |
| Deletion | Chr1 | 6712671 | 6712675 | 4 | LOC_Os01g12290 |
| Deletion | Chr8 | 6819863 | 6819873 | 10 | LOC_Os08g11620 |
| Deletion | Chr6 | 7546035 | 7546036 | 1 | LOC_Os06g13630 |
| Deletion | Chr3 | 7619219 | 7619220 | 1 | |
| Deletion | Chr12 | 8281938 | 8281942 | 4 | |
| Deletion | Chr12 | 8893563 | 8893564 | 1 | |
| Deletion | Chr10 | 9298834 | 9298835 | 1 | LOC_Os10g18294 |
| Deletion | Chr7 | 9610637 | 9610650 | 13 | |
| Deletion | Chr1 | 9898681 | 9898683 | 2 | |
| Deletion | Chr2 | 10562542 | 10562556 | 14 | |
| Deletion | Chr9 | 11331683 | 11331689 | 6 | |
| Deletion | Chr11 | 15531553 | 15531556 | 3 | LOC_Os11g26990 |
| Deletion | Chr2 | 15725977 | 15725978 | 1 | |
| Deletion | Chr12 | 15792948 | 15792964 | 16 | |
| Deletion | Chr8 | 17818069 | 17818070 | 1 | |
| Deletion | Chr11 | 18050966 | 18050967 | 1 | |
| Deletion | Chr8 | 18553674 | 18563755 | 10081 | 3 |
| Deletion | Chr11 | 18679639 | 18679696 | 57 | |
| Deletion | Chr9 | 19548665 | 19548673 | 8 | |
| Deletion | Chr1 | 21829282 | 21829283 | 1 | |
| Deletion | Chr2 | 22179031 | 22179048 | 17 | |
| Deletion | Chr2 | 22532370 | 22532371 | 1 | |
| Deletion | Chr10 | 23135804 | 23135806 | 2 | |
| Deletion | Chr2 | 23533779 | 23533782 | 3 | |
| Deletion | Chr1 | 25965401 | 25965404 | 3 | |
| Deletion | Chr1 | 25991001 | 26077000 | 86000 | 14 |
| Deletion | Chr11 | 26803624 | 26803647 | 23 | |
| Deletion | Chr7 | 27513761 | 27513764 | 3 | |
| Deletion | Chr4 | 27897600 | 27897620 | 20 | |
| Deletion | Chr4 | 28192256 | 28192257 | 1 | LOC_Os04g47530 |
| Deletion | Chr1 | 28872373 | 28874279 | 1906 | LOC_Os01g50259 |
| Deletion | Chr4 | 30951979 | 30951981 | 2 | LOC_Os04g52110 |
| Deletion | Chr4 | 33033011 | 33044918 | 11907 | 2 |
| Deletion | Chr1 | 34200046 | 34200048 | 2 | |
| Deletion | Chr1 | 42574602 | 42574608 | 6 |
Insertions: 12
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 5037963 | 6933453 | |
| Inversion | Chr7 | 11686342 | 11745117 | LOC_Os07g20270 |
| Inversion | Chr10 | 13197353 | 16207573 | LOC_Os10g31030 |
| Inversion | Chr5 | 19097731 | 20289393 | LOC_Os05g32610 |
| Inversion | Chr1 | 25991732 | 27264933 | LOC_Os01g47650 |
| Inversion | Chr1 | 26002474 | 27264838 | LOC_Os01g47650 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 993875 | Chr1 | 28874273 | LOC_Os09g02400 |
| Translocation | Chr9 | 993882 | Chr1 | 28872373 | 2 |
| Translocation | Chr9 | 2717453 | Chr7 | 24948461 | LOC_Os07g41610 |
| Translocation | Chr10 | 9149132 | Chr1 | 26012073 | |
| Translocation | Chr12 | 13386894 | Chr6 | 13714514 | LOC_Os06g23504 |
| Translocation | Chr9 | 15061219 | Chr4 | 29266248 | LOC_Os09g25130 |
| Translocation | Chr12 | 18797982 | Chr3 | 20578516 | LOC_Os03g37130 |
| Translocation | Chr11 | 21132545 | Chr7 | 11686355 | LOC_Os07g20270 |
| Translocation | Chr9 | 21144747 | Chr1 | 25991766 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr2 | 16294367 | 16294367 | 0 | LOC_Os02g27520 |
