Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1610-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1610-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 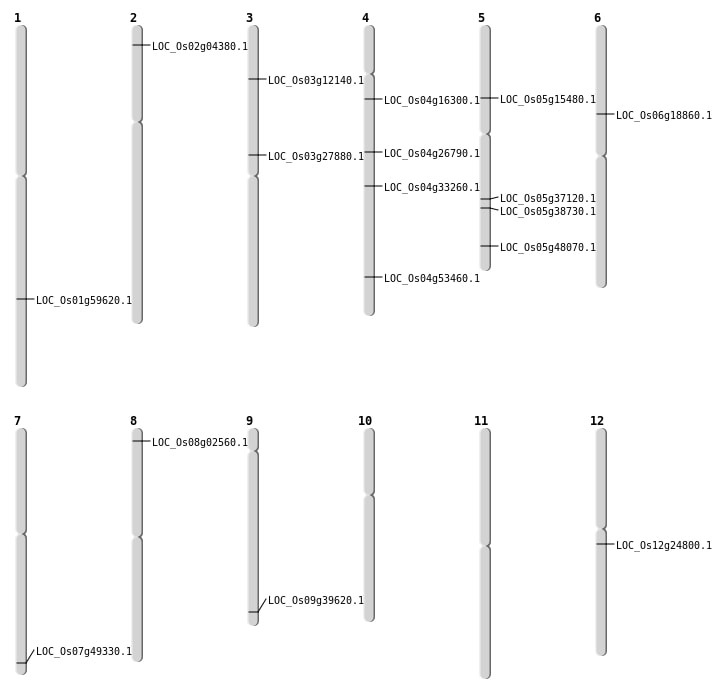
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10696927 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 14709059 | C-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr1 | 34483389 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g59620 |
| SBS | Chr1 | 41178235 | T-C | Homo | INTERGENIC | |
| SBS | Chr1 | 6180108 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 7401094 | A-C | HET | INTRON | |
| SBS | Chr10 | 1895170 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 18720893 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 27791618 | A-T | Homo | INTRON | |
| SBS | Chr12 | 14233560 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g24800 |
| SBS | Chr12 | 8988795 | T-C | HET | INTRON | |
| SBS | Chr2 | 1931633 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g04380 |
| SBS | Chr2 | 2622349 | A-C | HET | INTRON | |
| SBS | Chr2 | 31103862 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 6520622 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 16998640 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28900364 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 28900365 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 35589201 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 6374528 | A-C | HET | INTERGENIC | |
| SBS | Chr5 | 21694181 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 21694182 | C-T | HET | STOP_GAINED | LOC_Os05g37120 |
| SBS | Chr5 | 24494094 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24896861 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 8755189 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15480 |
| SBS | Chr6 | 24180715 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr7 | 13029351 | A-G | HET | INTRON | |
| SBS | Chr7 | 22710315 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 3856976 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 1056674 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g02560 |
| SBS | Chr8 | 19950223 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 7516616 | T-C | Homo | INTERGENIC | |
| SBS | Chr9 | 14135354 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 18162875 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 22594118 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 5918410 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 7089383 | C-T | HET | INTERGENIC |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 384872 | 384876 | 4 | |
| Deletion | Chr9 | 1924060 | 1924078 | 18 | |
| Deletion | Chr4 | 3011065 | 3011066 | 1 | |
| Deletion | Chr11 | 4505983 | 4505984 | 1 | |
| Deletion | Chr3 | 6372746 | 6372752 | 6 | LOC_Os03g12140 |
| Deletion | Chr7 | 10528398 | 10528399 | 1 | |
| Deletion | Chr10 | 10710556 | 10710560 | 4 | |
| Deletion | Chr6 | 10716561 | 10716572 | 11 | LOC_Os06g18860 |
| Deletion | Chr1 | 11075483 | 11075484 | 1 | |
| Deletion | Chr9 | 13056362 | 13056364 | 2 | |
| Deletion | Chr3 | 13414962 | 13414963 | 1 | |
| Deletion | Chr8 | 13517881 | 13517882 | 1 | |
| Deletion | Chr6 | 13632335 | 13632336 | 1 | |
| Deletion | Chr3 | 14905933 | 14905943 | 10 | |
| Deletion | Chr4 | 15637306 | 15666566 | 29260 | LOC_Os04g26790 |
| Deletion | Chr7 | 15779408 | 15779409 | 1 | |
| Deletion | Chr3 | 16018717 | 16018723 | 6 | LOC_Os03g27880 |
| Deletion | Chr1 | 16454057 | 16454066 | 9 | |
| Deletion | Chr4 | 20121862 | 20121863 | 1 | LOC_Os04g33260 |
| Deletion | Chr2 | 21121839 | 21121870 | 31 | |
| Deletion | Chr3 | 21957000 | 21957006 | 6 | |
| Deletion | Chr5 | 22730572 | 22730596 | 24 | LOC_Os05g38730 |
| Deletion | Chr9 | 22737770 | 22737815 | 45 | LOC_Os09g39620 |
| Deletion | Chr8 | 24651115 | 24651121 | 6 | |
| Deletion | Chr2 | 25427761 | 25427767 | 6 | |
| Deletion | Chr11 | 27703560 | 27703562 | 2 | |
| Deletion | Chr7 | 29548168 | 29548169 | 1 | LOC_Os07g49330 |
| Deletion | Chr4 | 31381926 | 31381939 | 13 | |
| Deletion | Chr3 | 33407445 | 33407457 | 12 |
Insertions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 41514524 | 41514524 | 1 | |
| Insertion | Chr1 | 48990 | 48990 | 1 | |
| Insertion | Chr1 | 7754757 | 7754757 | 1 | |
| Insertion | Chr11 | 13994166 | 13994166 | 1 | |
| Insertion | Chr11 | 7308109 | 7308111 | 3 | |
| Insertion | Chr12 | 14481154 | 14481154 | 1 | |
| Insertion | Chr12 | 15024484 | 15024496 | 13 | |
| Insertion | Chr3 | 30494350 | 30494350 | 1 | |
| Insertion | Chr4 | 16434830 | 16434835 | 6 | |
| Insertion | Chr4 | 23106402 | 23106402 | 1 | |
| Insertion | Chr5 | 27560817 | 27560820 | 4 | LOC_Os05g48070 |
| Insertion | Chr5 | 6018131 | 6018131 | 1 | |
| Insertion | Chr7 | 13461159 | 13461159 | 1 | |
| Insertion | Chr7 | 23389083 | 23389109 | 27 | |
| Insertion | Chr7 | 24289012 | 24289013 | 2 | |
| Insertion | Chr7 | 8338880 | 8338881 | 2 | |
| Insertion | Chr8 | 7862935 | 7862935 | 1 | |
| Insertion | Chr8 | 9212042 | 9212042 | 1 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 31543412 | 31833567 | LOC_Os04g53460 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 2570542 | Chr1 | 8218341 | |
| Translocation | Chr2 | 18066728 | Chr1 | 8221598 | |
| Translocation | Chr10 | 18195122 | Chr4 | 8855403 | LOC_Os04g16300 |
| Translocation | Chr8 | 25425880 | Chr1 | 34371005 |
