Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1614-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1614-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 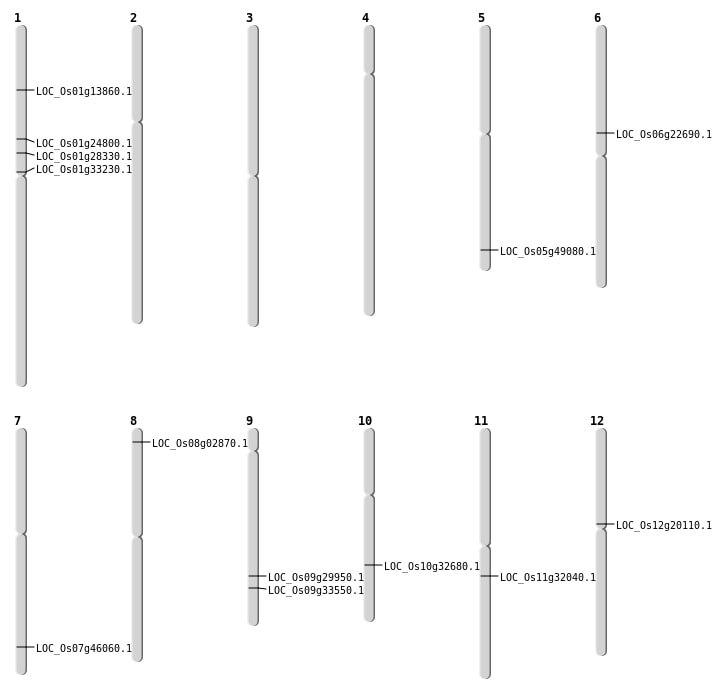
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13233497 | A-G | HET | INTRON | |
| SBS | Chr1 | 41896095 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr10 | 17113025 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g32680 |
| SBS | Chr11 | 14235749 | G-T | Homo | INTERGENIC | |
| SBS | Chr11 | 14869474 | A-T | Homo | INTERGENIC | |
| SBS | Chr11 | 24895442 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 5998009 | T-C | Homo | INTERGENIC | |
| SBS | Chr12 | 11693410 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g20110 |
| SBS | Chr12 | 22764728 | G-T | Homo | INTRON | |
| SBS | Chr12 | 978163 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 4926268 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 10078598 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 12039639 | G-T | Homo | INTERGENIC | |
| SBS | Chr4 | 24583998 | A-T | Homo | INTRON | |
| SBS | Chr4 | 7694307 | A-G | Homo | SYNONYMOUS_CODING | |
| SBS | Chr5 | 15109742 | A-T | HET | INTERGENIC | |
| SBS | Chr5 | 26174777 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 6541612 | A-G | HET | UTR_3_PRIME | |
| SBS | Chr5 | 8273001 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 13061083 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 14237915 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 11251124 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 11536855 | G-C | HET | INTRON | |
| SBS | Chr7 | 1427692 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 27488890 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g46060 |
| SBS | Chr8 | 11833374 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 11833375 | T-C | Homo | INTERGENIC | |
| SBS | Chr8 | 13654777 | T-G | Homo | INTERGENIC | |
| SBS | Chr8 | 4412467 | C-T | Homo | INTRON | |
| SBS | Chr9 | 20211682 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 22748715 | T-C | Homo | INTRON |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 656835 | 656837 | 2 | |
| Deletion | Chr4 | 1612052 | 1612053 | 1 | |
| Deletion | Chr1 | 7773093 | 7773099 | 6 | LOC_Os01g13860 |
| Deletion | Chr8 | 10600626 | 10600627 | 1 | |
| Deletion | Chr8 | 11636721 | 11636726 | 5 | |
| Deletion | Chr6 | 13196928 | 13196935 | 7 | LOC_Os06g22690 |
| Deletion | Chr1 | 13970112 | 13970115 | 3 | LOC_Os01g24800 |
| Deletion | Chr7 | 14247041 | 14247047 | 6 | |
| Deletion | Chr10 | 15519580 | 15519590 | 10 | |
| Deletion | Chr1 | 15869500 | 15869503 | 3 | LOC_Os01g28330 |
| Deletion | Chr11 | 16452630 | 16452631 | 1 | |
| Deletion | Chr1 | 17767554 | 17767556 | 2 | |
| Deletion | Chr4 | 18239290 | 18244753 | 5463 | |
| Deletion | Chr1 | 18298299 | 18298308 | 9 | LOC_Os01g33230 |
| Deletion | Chr4 | 18541475 | 18541482 | 7 | |
| Deletion | Chr11 | 18883631 | 18883637 | 6 | LOC_Os11g32040 |
| Deletion | Chr9 | 19783750 | 19783751 | 1 | LOC_Os09g33550 |
| Deletion | Chr11 | 20475812 | 20475813 | 1 | |
| Deletion | Chr6 | 21962018 | 21962019 | 1 | |
| Deletion | Chr5 | 28133448 | 28133449 | 1 | LOC_Os05g49080 |
| Deletion | Chr2 | 31376553 | 31376555 | 2 | |
| Deletion | Chr2 | 32109917 | 32109918 | 1 |
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 11406649 | 11406649 | 1 | |
| Insertion | Chr11 | 8055179 | 8055219 | 41 | |
| Insertion | Chr3 | 11804029 | 11804029 | 1 | |
| Insertion | Chr4 | 25120888 | 25120905 | 18 | |
| Insertion | Chr4 | 30673104 | 30673105 | 2 | |
| Insertion | Chr4 | 8691608 | 8691608 | 1 | |
| Insertion | Chr5 | 13584506 | 13584506 | 1 | |
| Insertion | Chr5 | 8171042 | 8171042 | 1 | |
| Insertion | Chr6 | 25368332 | 25368332 | 1 | |
| Insertion | Chr6 | 25488478 | 25488478 | 1 | |
| Insertion | Chr7 | 25308283 | 25308328 | 46 | |
| Insertion | Chr9 | 18225801 | 18225801 | 1 | LOC_Os09g29950 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 16913410 | 16949649 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 9247513 | Chr2 | 16194708 | |
| Translocation | Chr6 | 10144734 | Chr1 | 1692522 | |
| Translocation | Chr10 | 10311980 | Chr4 | 5525937 | |
| Translocation | Chr7 | 26712210 | Chr4 | 33521210 | |
| Translocation | Chr11 | 26925422 | Chr8 | 1219480 | LOC_Os08g02870 |
