Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1618-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1618-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 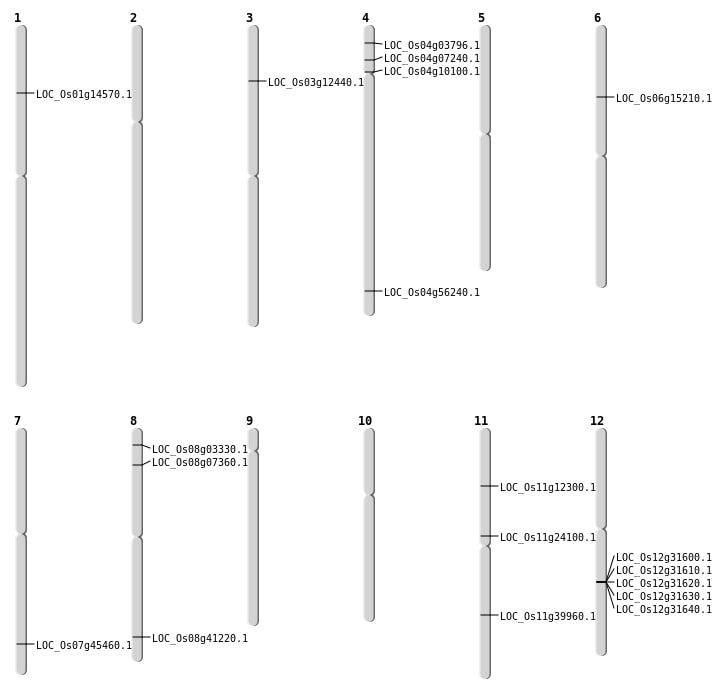
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 25857315 | C-A | HET | INTERGENIC | |
| SBS | Chr1 | 35011677 | C-G | HET | INTERGENIC | |
| SBS | Chr1 | 39341571 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 24237317 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 1616462 | C-G | Homo | INTERGENIC | |
| SBS | Chr2 | 22315533 | G-C | HET | INTERGENIC | |
| SBS | Chr2 | 25121400 | A-C | HET | INTRON | |
| SBS | Chr3 | 14413091 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 17545 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 3842322 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g07240 |
| SBS | Chr4 | 4634355 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 5450467 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g10100 |
| SBS | Chr4 | 7707189 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 7736339 | G-A | HET | INTRON | |
| SBS | Chr4 | 8128495 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 27855170 | G-C | HET | INTERGENIC | |
| SBS | Chr5 | 634074 | G-A | HET | INTRON | |
| SBS | Chr6 | 10359719 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 15114125 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 17695048 | T-C | HET | INTERGENIC | |
| SBS | Chr6 | 9689571 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 1544448 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g03330 |
| SBS | Chr8 | 26029643 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g41220 |
| SBS | Chr8 | 4118833 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g07360 |
| SBS | Chr9 | 5886411 | C-A | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 836518 | 836519 | 1 | |
| Deletion | Chr4 | 1514932 | 1514944 | 12 | |
| Deletion | Chr12 | 1661339 | 1661340 | 1 | |
| Deletion | Chr4 | 1710222 | 1710233 | 11 | LOC_Os04g03796 |
| Deletion | Chr10 | 2285296 | 2287837 | 2541 | |
| Deletion | Chr8 | 5051065 | 5051075 | 10 | |
| Deletion | Chr10 | 5393401 | 5393406 | 5 | |
| Deletion | Chr10 | 5565396 | 5565402 | 6 | |
| Deletion | Chr1 | 5836653 | 5836656 | 3 | |
| Deletion | Chr3 | 6568042 | 6568048 | 6 | LOC_Os03g12440 |
| Deletion | Chr11 | 6872965 | 6872969 | 4 | LOC_Os11g12300 |
| Deletion | Chr12 | 7401501 | 7401502 | 1 | |
| Deletion | Chr2 | 7689975 | 7689976 | 1 | |
| Deletion | Chr4 | 8128496 | 8128501 | 5 | |
| Deletion | Chr1 | 8166631 | 8166634 | 3 | LOC_Os01g14570 |
| Deletion | Chr7 | 8984896 | 8984897 | 1 | |
| Deletion | Chr11 | 13700225 | 13700226 | 1 | LOC_Os11g24100 |
| Deletion | Chr3 | 16716078 | 16716095 | 17 | |
| Deletion | Chr8 | 16922091 | 16922092 | 1 | |
| Deletion | Chr5 | 17571294 | 17571303 | 9 | |
| Deletion | Chr9 | 17667030 | 17667032 | 2 | |
| Deletion | Chr12 | 19019001 | 19056000 | 37000 | 5 |
| Deletion | Chr8 | 19655096 | 19655098 | 2 | |
| Deletion | Chr5 | 21506750 | 21506752 | 2 | |
| Deletion | Chr11 | 22254354 | 22254366 | 12 | |
| Deletion | Chr9 | 22641664 | 22641678 | 14 | |
| Deletion | Chr11 | 23819925 | 23819933 | 8 | LOC_Os11g39960 |
| Deletion | Chr12 | 25231021 | 25231022 | 1 | |
| Deletion | Chr7 | 27122135 | 27122137 | 2 | LOC_Os07g45460 |
| Deletion | Chr4 | 27547034 | 27547038 | 4 | |
| Deletion | Chr3 | 28536164 | 28536177 | 13 | |
| Deletion | Chr2 | 30240761 | 30240762 | 1 | |
| Deletion | Chr4 | 32655245 | 32655248 | 3 | |
| Deletion | Chr3 | 32796808 | 32796809 | 1 | |
| Deletion | Chr3 | 34965807 | 34965811 | 4 | |
| Deletion | Chr1 | 40449170 | 40449174 | 4 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17809437 | 17809440 | 4 | |
| Insertion | Chr1 | 30072306 | 30072365 | 60 | |
| Insertion | Chr10 | 13688979 | 13688980 | 2 | |
| Insertion | Chr10 | 3986710 | 3986711 | 2 | |
| Insertion | Chr11 | 10895601 | 10895601 | 1 | |
| Insertion | Chr11 | 11895259 | 11895262 | 4 | |
| Insertion | Chr2 | 6766927 | 6766930 | 4 | |
| Insertion | Chr3 | 32604039 | 32604039 | 1 | |
| Insertion | Chr4 | 33165043 | 33165066 | 24 | |
| Insertion | Chr6 | 17019334 | 17019334 | 1 | |
| Insertion | Chr6 | 8619234 | 8619255 | 22 | LOC_Os06g15210 |
| Insertion | Chr7 | 26107125 | 26107126 | 2 | |
| Insertion | Chr8 | 17940641 | 17940641 | 1 | |
| Insertion | Chr9 | 1716835 | 1716837 | 3 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 19407676 | Chr1 | 27650033 | |
| Translocation | Chr4 | 33532190 | Chr1 | 41587107 | LOC_Os04g56240 |
