Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1619-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1619-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 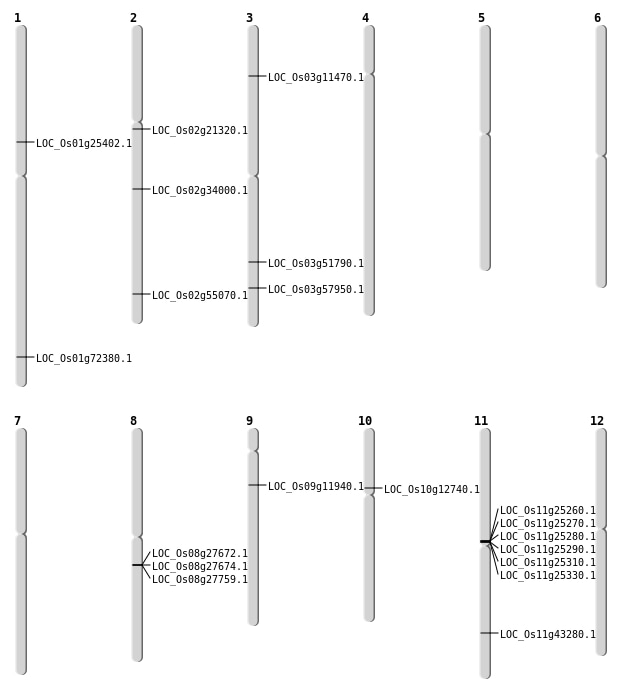
|
| Variant Information |
|---|
Single base substitutions: 44
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21079068 | C-A | Homo | INTRON | |
| SBS | Chr1 | 32439563 | G-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 36036711 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 8742485 | T-C | Homo | INTERGENIC | |
| SBS | Chr10 | 17834058 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 5230666 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 7103540 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g12740 |
| SBS | Chr11 | 10061702 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 26111212 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os11g43280 |
| SBS | Chr11 | 7391556 | G-T | Homo | INTERGENIC | |
| SBS | Chr12 | 10844427 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 25155415 | G-T | Homo | INTERGENIC | |
| SBS | Chr12 | 8951291 | C-T | HET | INTRON | |
| SBS | Chr2 | 10458660 | G-T | Homo | INTERGENIC | |
| SBS | Chr2 | 23962042 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 33733631 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g55070 |
| SBS | Chr2 | 899098 | C-G | HET | SPLICE_SITE_REGION | |
| SBS | Chr3 | 11076762 | C-T | HET | INTRON | |
| SBS | Chr3 | 30903703 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 33122697 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 5922104 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g11470 |
| SBS | Chr4 | 812731 | G-T | Homo | INTERGENIC | |
| SBS | Chr5 | 27210257 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 28768045 | T-A | HET | INTRON | |
| SBS | Chr5 | 29416887 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr5 | 4052628 | G-T | HET | INTERGENIC | |
| SBS | Chr6 | 11469000 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 13083044 | A-T | HET | INTERGENIC | |
| SBS | Chr6 | 20218978 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 26151149 | C-T | HET | INTERGENIC | |
| SBS | Chr6 | 8448560 | T-C | HET | SPLICE_SITE_REGION | |
| SBS | Chr7 | 10608673 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 16253726 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 17265759 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 25283948 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 29253604 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 9444354 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 10825215 | T-C | HET | INTRON | |
| SBS | Chr8 | 17455807 | T-A | HET | INTRON | |
| SBS | Chr8 | 23147055 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 3804896 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 6722121 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g11940 |
| SBS | Chr9 | 8873948 | G-T | Homo | INTERGENIC | |
| SBS | Chr9 | 9147209 | G-A | HET | INTRON |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 2881237 | 2881247 | 10 | |
| Deletion | Chr5 | 3060231 | 3060232 | 1 | |
| Deletion | Chr12 | 5599215 | 5599342 | 127 | |
| Deletion | Chr5 | 9039406 | 9039412 | 6 | |
| Deletion | Chr2 | 9579989 | 9580008 | 19 | |
| Deletion | Chr2 | 9588487 | 9588506 | 19 | |
| Deletion | Chr12 | 9979371 | 9979372 | 1 | |
| Deletion | Chr2 | 10406190 | 10406191 | 1 | |
| Deletion | Chr4 | 10644497 | 10644503 | 6 | |
| Deletion | Chr8 | 10717865 | 10717868 | 3 | |
| Deletion | Chr2 | 12654030 | 12654043 | 13 | LOC_Os02g21320 |
| Deletion | Chr11 | 14391001 | 14429000 | 38000 | 6 |
| Deletion | Chr2 | 16762902 | 16762910 | 8 | |
| Deletion | Chr8 | 16863499 | 16863516 | 17 | 2 |
| Deletion | Chr8 | 16912515 | 16912551 | 36 | LOC_Os08g27759 |
| Deletion | Chr8 | 17852926 | 17852929 | 3 | |
| Deletion | Chr3 | 18222230 | 18222240 | 10 | |
| Deletion | Chr10 | 18899571 | 18899572 | 1 | |
| Deletion | Chr7 | 19026030 | 19026032 | 2 | |
| Deletion | Chr12 | 19688815 | 19688816 | 1 | |
| Deletion | Chr5 | 20117218 | 20117219 | 1 | |
| Deletion | Chr2 | 20297036 | 20297037 | 1 | LOC_Os02g34000 |
| Deletion | Chr8 | 21164568 | 21164597 | 29 | |
| Deletion | Chr10 | 22287066 | 22287067 | 1 | |
| Deletion | Chr2 | 22612125 | 22612127 | 2 | |
| Deletion | Chr6 | 23101253 | 23101257 | 4 | |
| Deletion | Chr7 | 25468869 | 25468885 | 16 | |
| Deletion | Chr12 | 25835322 | 25835335 | 13 | |
| Deletion | Chr6 | 25929896 | 25929897 | 1 | |
| Deletion | Chr5 | 27628253 | 27628274 | 21 | |
| Deletion | Chr5 | 28756190 | 28756191 | 1 | |
| Deletion | Chr3 | 29706275 | 29706276 | 1 | LOC_Os03g51790 |
| Deletion | Chr1 | 32697667 | 32697678 | 11 | |
| Deletion | Chr3 | 33012916 | 33012917 | 1 | LOC_Os03g57950 |
| Deletion | Chr3 | 33197094 | 33197105 | 11 | |
| Deletion | Chr4 | 34225539 | 34225541 | 2 | |
| Deletion | Chr1 | 41983468 | 41983469 | 1 | LOC_Os01g72380 |
Insertions: 13
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 14374246 | 14899593 | LOC_Os01g25402 |
| Inversion | Chr1 | 14732573 | 15964804 | LOC_Os01g25990 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 15550226 | Chr5 | 5643363 | |
| Translocation | Chr8 | 18930324 | Chr1 | 3645085 |
