Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1622-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1622-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 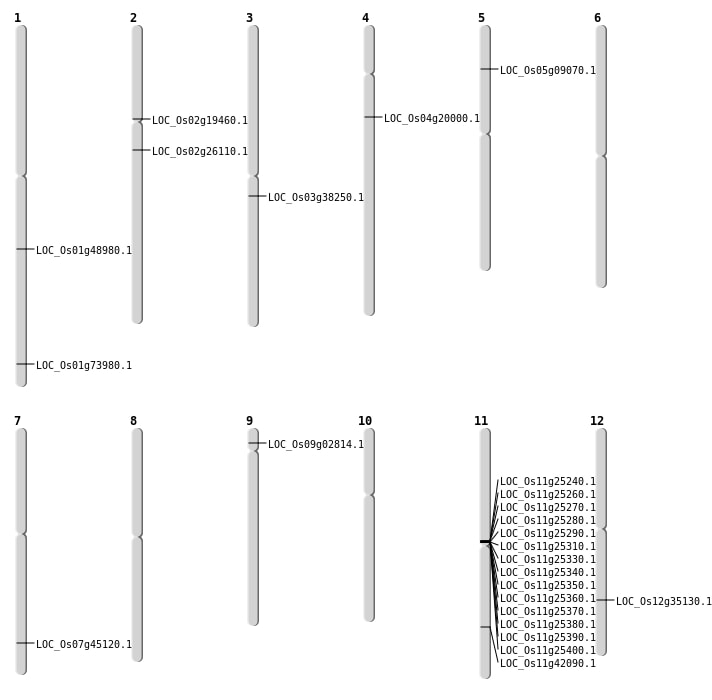
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13177848 | G-T | Homo | INTERGENIC | |
| SBS | Chr1 | 23570377 | A-T | Homo | INTRON | |
| SBS | Chr1 | 23570379 | G-T | Homo | INTRON | |
| SBS | Chr1 | 25603361 | C-A | Homo | INTERGENIC | |
| SBS | Chr1 | 5642793 | C-A | HET | INTERGENIC | |
| SBS | Chr10 | 15455237 | A-G | HET | INTERGENIC | |
| SBS | Chr10 | 23031584 | A-G | Homo | UTR_3_PRIME | |
| SBS | Chr11 | 14358174 | C-G | HET | INTERGENIC | |
| SBS | Chr11 | 22084163 | G-C | HET | INTERGENIC | |
| SBS | Chr11 | 25344579 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g42090 |
| SBS | Chr11 | 27995258 | A-T | HET | INTRON | |
| SBS | Chr12 | 21377748 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35130 |
| SBS | Chr12 | 3609418 | C-T | HET | INTERGENIC | |
| SBS | Chr2 | 15343009 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g26110 |
| SBS | Chr3 | 6252121 | C-G | HET | INTERGENIC | |
| SBS | Chr4 | 10679321 | A-G | Homo | INTERGENIC | |
| SBS | Chr4 | 23095495 | C-A | Homo | INTRON | |
| SBS | Chr4 | 2955037 | A-G | HET | INTERGENIC | |
| SBS | Chr5 | 22078482 | C-T | HET | INTRON | |
| SBS | Chr5 | 26483046 | G-C | HET | INTERGENIC | |
| SBS | Chr6 | 17280587 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 23713889 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 5727061 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 20249343 | T-C | Homo | UTR_5_PRIME | |
| SBS | Chr9 | 17889772 | C-T | HET | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1317241 | 1317242 | 1 | LOC_Os09g02814 |
| Deletion | Chr6 | 2233332 | 2233334 | 2 | |
| Deletion | Chr11 | 2295243 | 2295247 | 4 | |
| Deletion | Chr11 | 2637171 | 2637172 | 1 | |
| Deletion | Chr12 | 3528541 | 3528547 | 6 | |
| Deletion | Chr10 | 4696510 | 4696534 | 24 | |
| Deletion | Chr2 | 7168866 | 7168870 | 4 | |
| Deletion | Chr9 | 8183982 | 8183988 | 6 | |
| Deletion | Chr12 | 8735351 | 8735352 | 1 | |
| Deletion | Chr6 | 11430463 | 11430471 | 8 | |
| Deletion | Chr7 | 12258334 | 12264235 | 5901 | |
| Deletion | Chr1 | 13124619 | 13124627 | 8 | |
| Deletion | Chr11 | 14376001 | 14483000 | 107000 | 14 |
| Deletion | Chr2 | 17318818 | 17318827 | 9 | |
| Deletion | Chr3 | 21234872 | 21234873 | 1 | LOC_Os03g38250 |
| Deletion | Chr11 | 22308787 | 22308798 | 11 | |
| Deletion | Chr2 | 23869770 | 23869776 | 6 | |
| Deletion | Chr5 | 24476906 | 24476933 | 27 | |
| Deletion | Chr2 | 24654012 | 24654017 | 5 | |
| Deletion | Chr3 | 24741462 | 24741472 | 10 | |
| Deletion | Chr1 | 26771675 | 26771676 | 1 | |
| Deletion | Chr1 | 27040791 | 27040792 | 1 | |
| Deletion | Chr5 | 27904717 | 27904721 | 4 | |
| Deletion | Chr6 | 30195443 | 30195445 | 2 | |
| Deletion | Chr2 | 30357925 | 30357937 | 12 | |
| Deletion | Chr1 | 42857447 | 42857451 | 4 | LOC_Os01g73980 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 13350424 | 13350444 | 21 | |
| Insertion | Chr1 | 13554297 | 13554298 | 2 | |
| Insertion | Chr4 | 8260967 | 8260973 | 7 | |
| Insertion | Chr5 | 5037856 | 5037914 | 59 | LOC_Os05g09070 |
| Insertion | Chr6 | 24944786 | 24944786 | 1 | |
| Insertion | Chr7 | 4420152 | 4420163 | 12 | |
| Insertion | Chr8 | 13728309 | 13728311 | 3 | |
| Insertion | Chr8 | 4719724 | 4719803 | 80 | |
| Insertion | Chr9 | 9371299 | 9371349 | 51 |
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 615718 | 9357428 | |
| Inversion | Chr4 | 11150984 | 21743890 | LOC_Os04g20000 |
| Inversion | Chr11 | 14375979 | 14533828 | |
| Inversion | Chr11 | 14483661 | 14533826 | LOC_Os11g25400 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2299641 | Chr7 | 24482767 | |
| Translocation | Chr4 | 11150983 | Chr2 | 11365279 | LOC_Os04g20000 |
| Translocation | Chr4 | 11304027 | Chr2 | 11369346 | LOC_Os02g19460 |
| Translocation | Chr11 | 11899767 | Chr3 | 33248227 | |
| Translocation | Chr12 | 13389330 | Chr1 | 28118122 | LOC_Os01g48980 |
| Translocation | Chr3 | 17136827 | Chr2 | 24543549 | |
| Translocation | Chr3 | 20789164 | Chr1 | 4653367 | |
| Translocation | Chr7 | 26937037 | Chr5 | 14654919 | LOC_Os07g45120 |
