Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1623-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1623-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 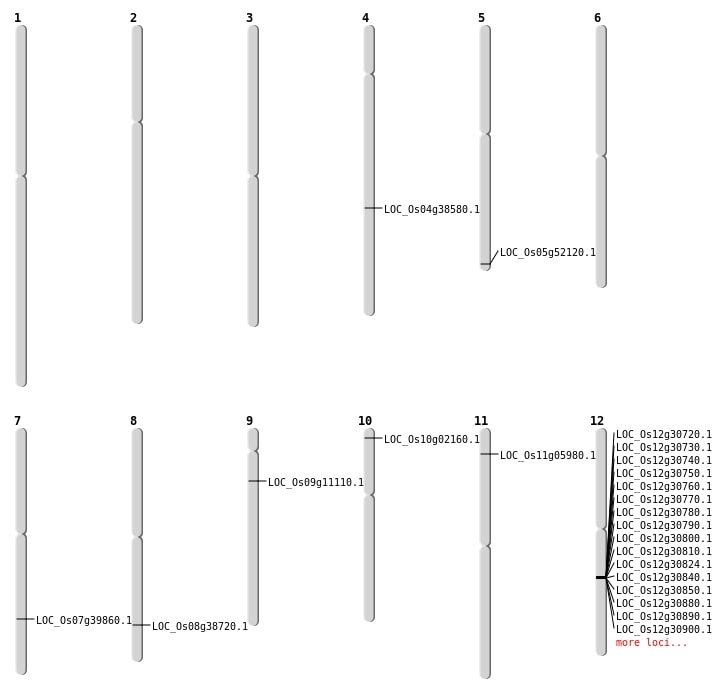
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22972324 | T-A | Homo | INTERGENIC | |
| SBS | Chr1 | 28566540 | A-T | Homo | INTERGENIC | |
| SBS | Chr1 | 38743929 | C-T | Homo | INTRON | |
| SBS | Chr1 | 40685653 | T-A | Homo | INTERGENIC | |
| SBS | Chr10 | 2141532 | A-T | Homo | INTERGENIC | |
| SBS | Chr10 | 5486005 | T-A | Homo | INTERGENIC | |
| SBS | Chr10 | 8080302 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 20548777 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 8794086 | G-C | HET | INTRON | |
| SBS | Chr12 | 12350668 | A-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 19509480 | G-T | HET | INTRON | |
| SBS | Chr12 | 8954890 | G-T | HET | INTRON | |
| SBS | Chr2 | 12426862 | T-C | Homo | INTERGENIC | |
| SBS | Chr3 | 11279323 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 22827488 | C-T | Homo | UTR_5_PRIME | |
| SBS | Chr3 | 34375686 | A-C | HET | INTERGENIC | |
| SBS | Chr3 | 35311212 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 10792584 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr4 | 1538725 | C-T | Homo | INTERGENIC | |
| SBS | Chr4 | 2352407 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20865004 | A-T | Homo | INTRON | |
| SBS | Chr6 | 21338836 | C-T | Homo | INTERGENIC | |
| SBS | Chr6 | 27922060 | C-G | HET | INTERGENIC | |
| SBS | Chr6 | 4392482 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14026266 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 1810383 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 183229 | A-C | Homo | UTR_5_PRIME | |
| SBS | Chr7 | 183231 | G-A | Homo | UTR_5_PRIME | |
| SBS | Chr7 | 5873087 | A-T | HET | INTERGENIC | |
| SBS | Chr8 | 11789663 | G-A | Homo | INTRON | |
| SBS | Chr8 | 20680551 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 25162059 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 19457722 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 4067243 | C-A | Homo | INTRON | |
| SBS | Chr9 | 6162278 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g11110 |
| SBS | Chr9 | 9237013 | A-G | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 606451 | 606452 | 1 | |
| Deletion | Chr4 | 609741 | 609742 | 1 | |
| Deletion | Chr10 | 736883 | 736890 | 7 | LOC_Os10g02160 |
| Deletion | Chr9 | 1152149 | 1152154 | 5 | |
| Deletion | Chr8 | 1431220 | 1431228 | 8 | |
| Deletion | Chr8 | 1440131 | 1440139 | 8 | |
| Deletion | Chr9 | 2680241 | 2680246 | 5 | |
| Deletion | Chr11 | 2815547 | 2815549 | 2 | LOC_Os11g05980 |
| Deletion | Chr6 | 4407128 | 4407134 | 6 | |
| Deletion | Chr3 | 10366424 | 10366427 | 3 | |
| Deletion | Chr4 | 11410885 | 11410893 | 8 | |
| Deletion | Chr10 | 11962293 | 11965982 | 3689 | |
| Deletion | Chr10 | 13563562 | 13563564 | 2 | |
| Deletion | Chr10 | 14353405 | 14353676 | 271 | |
| Deletion | Chr3 | 14415369 | 14415370 | 1 | |
| Deletion | Chr4 | 17371198 | 17371199 | 1 | |
| Deletion | Chr8 | 17643067 | 17643068 | 1 | |
| Deletion | Chr12 | 18450001 | 18584000 | 134000 | 19 |
| Deletion | Chr10 | 19880077 | 19880078 | 1 | |
| Deletion | Chr8 | 20228476 | 20228477 | 1 | |
| Deletion | Chr4 | 20574010 | 20574011 | 1 | |
| Deletion | Chr3 | 22594174 | 22594181 | 7 | |
| Deletion | Chr7 | 23903042 | 23903043 | 1 | LOC_Os07g39860 |
| Deletion | Chr6 | 24073330 | 24073340 | 10 | |
| Deletion | Chr8 | 24484905 | 24484907 | 2 | LOC_Os08g38720 |
| Deletion | Chr11 | 25185311 | 25185312 | 1 | |
| Deletion | Chr6 | 25342701 | 25342704 | 3 | |
| Deletion | Chr5 | 25428218 | 25428219 | 1 | |
| Deletion | Chr6 | 26102884 | 26102887 | 3 | |
| Deletion | Chr3 | 27884090 | 27884092 | 2 | |
| Deletion | Chr5 | 29933161 | 29957175 | 24014 | LOC_Os05g52120 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 16406461 | 16406461 | 1 | |
| Insertion | Chr10 | 13451873 | 13451911 | 39 | |
| Insertion | Chr10 | 18094826 | 18094826 | 1 | |
| Insertion | Chr10 | 8216531 | 8216531 | 1 | |
| Insertion | Chr11 | 20877812 | 20877855 | 44 | |
| Insertion | Chr11 | 9843451 | 9843451 | 1 | |
| Insertion | Chr12 | 18512029 | 18512080 | 52 | |
| Insertion | Chr4 | 22925722 | 22925743 | 22 | LOC_Os04g38580 |
| Insertion | Chr4 | 29205757 | 29205771 | 15 | |
| Insertion | Chr4 | 8411775 | 8411775 | 1 | |
| Insertion | Chr6 | 18791059 | 18791102 | 44 | |
| Insertion | Chr7 | 11426019 | 11426072 | 54 | |
| Insertion | Chr7 | 20208158 | 20208158 | 1 | |
| Insertion | Chr7 | 22426877 | 22426880 | 4 | |
| Insertion | Chr7 | 25872667 | 25872667 | 1 | |
| Insertion | Chr9 | 15159192 | 15159227 | 36 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 21266764 | 21815016 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 21983057 | Chr5 | 12725490 | |
| Translocation | Chr11 | 24622474 | Chr2 | 11417389 |
