Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1625-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1625-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 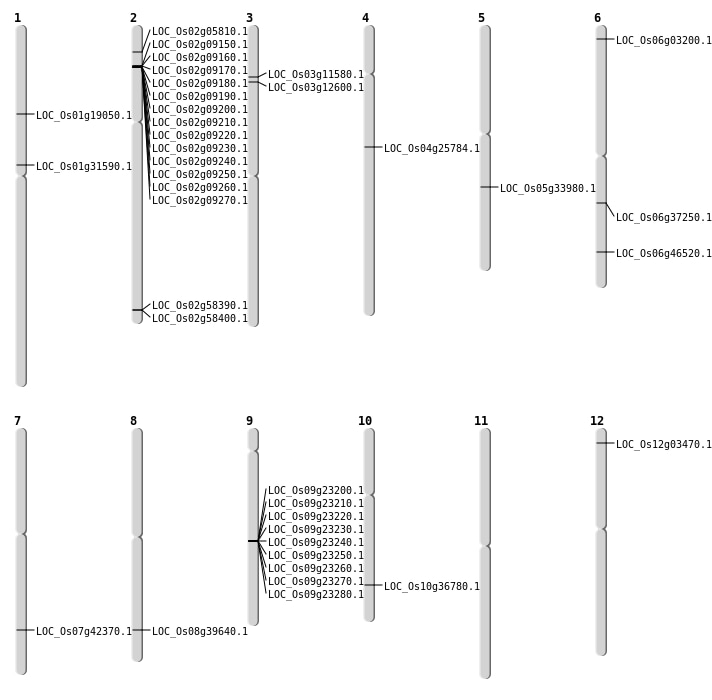
|
| Variant Information |
|---|
Single base substitutions: 43
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17287349 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g31590 |
| SBS | Chr1 | 20879771 | C-T | HET | INTRON | |
| SBS | Chr1 | 29472767 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 42356389 | G-C | Homo | INTERGENIC | |
| SBS | Chr1 | 43183354 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 6413835 | G-A | HET | INTRON | |
| SBS | Chr10 | 1029287 | C-T | HET | INTERGENIC | |
| SBS | Chr10 | 12212715 | C-T | Homo | INTERGENIC | |
| SBS | Chr10 | 19691925 | A-C | Homo | STOP_LOST | LOC_Os10g36780 |
| SBS | Chr10 | 5543226 | A-C | HET | UTR_5_PRIME | |
| SBS | Chr10 | 972685 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 1415562 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 1415563 | G-A | Homo | INTERGENIC | |
| SBS | Chr11 | 9896751 | A-G | Homo | UTR_3_PRIME | |
| SBS | Chr12 | 1093998 | A-T | HET | UTR_3_PRIME | |
| SBS | Chr12 | 5066445 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr12 | 7785851 | T-G | HET | INTERGENIC | |
| SBS | Chr2 | 22804361 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 31974734 | T-A | HET | INTERGENIC | |
| SBS | Chr2 | 8150934 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 6008766 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g11580 |
| SBS | Chr4 | 21765384 | G-A | Homo | INTERGENIC | |
| SBS | Chr5 | 11346490 | G-T | HET | INTRON | |
| SBS | Chr5 | 14030327 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 15109189 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 20636146 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 27786373 | C-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr6 | 23077422 | G-A | Homo | INTRON | |
| SBS | Chr6 | 27404425 | A-C | HET | INTERGENIC | |
| SBS | Chr6 | 27961286 | A-T | HET | INTRON | |
| SBS | Chr6 | 28248330 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g46520 |
| SBS | Chr6 | 4781945 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 22124902 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 29454628 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 25105200 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g39640 |
| SBS | Chr8 | 25153267 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 5215928 | A-G | HET | INTERGENIC | |
| SBS | Chr9 | 13978047 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 15568122 | C-A | HET | INTERGENIC | |
| SBS | Chr9 | 18593617 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 18593618 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 589319 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 6315964 | C-T | HET | INTERGENIC |
Deletions: 31
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 157655 | 157657 | 2 | |
| Deletion | Chr9 | 773921 | 773989 | 68 | |
| Deletion | Chr2 | 1807636 | 1807637 | 1 | |
| Deletion | Chr6 | 2554725 | 2554728 | 3 | |
| Deletion | Chr2 | 2867063 | 2867080 | 17 | LOC_Os02g05810 |
| Deletion | Chr1 | 4631519 | 4631521 | 2 | |
| Deletion | Chr2 | 4697001 | 4779000 | 82000 | 13 |
| Deletion | Chr3 | 6684672 | 6689522 | 4850 | LOC_Os03g12600 |
| Deletion | Chr9 | 9479975 | 9479977 | 2 | |
| Deletion | Chr1 | 10088658 | 10088665 | 7 | |
| Deletion | Chr9 | 13756001 | 13823000 | 67000 | 9 |
| Deletion | Chr3 | 14715603 | 14715604 | 1 | |
| Deletion | Chr3 | 14826325 | 14826339 | 14 | |
| Deletion | Chr1 | 14887086 | 14887104 | 18 | |
| Deletion | Chr9 | 15392087 | 15392090 | 3 | |
| Deletion | Chr1 | 15722010 | 15722016 | 6 | |
| Deletion | Chr8 | 15978684 | 15978693 | 9 | |
| Deletion | Chr1 | 16290790 | 16290791 | 1 | |
| Deletion | Chr8 | 16802771 | 16802772 | 1 | |
| Deletion | Chr11 | 17494659 | 17494779 | 120 | |
| Deletion | Chr9 | 17678287 | 17678288 | 1 | |
| Deletion | Chr9 | 18198710 | 18198717 | 7 | |
| Deletion | Chr5 | 20065180 | 20065283 | 103 | LOC_Os05g33980 |
| Deletion | Chr6 | 20444075 | 20444079 | 4 | |
| Deletion | Chr6 | 25287126 | 25287127 | 1 | |
| Deletion | Chr7 | 25348094 | 25348096 | 2 | LOC_Os07g42370 |
| Deletion | Chr8 | 27842126 | 27842128 | 2 | |
| Deletion | Chr2 | 32147319 | 32147337 | 18 | |
| Deletion | Chr3 | 34797086 | 34797097 | 11 | |
| Deletion | Chr2 | 34863839 | 34863851 | 12 | |
| Deletion | Chr2 | 35704552 | 35714599 | 10047 | 2 |
Insertions: 15
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 10777941 | 10777958 | 18 | LOC_Os01g19050 |
| Insertion | Chr1 | 11387051 | 11387053 | 3 | |
| Insertion | Chr1 | 24524866 | 24524869 | 4 | |
| Insertion | Chr1 | 26513763 | 26513766 | 4 | |
| Insertion | Chr1 | 38401727 | 38401727 | 1 | |
| Insertion | Chr12 | 3701937 | 3701970 | 34 | |
| Insertion | Chr2 | 12938779 | 12938788 | 10 | |
| Insertion | Chr2 | 9359898 | 9359899 | 2 | |
| Insertion | Chr3 | 18056814 | 18056814 | 1 | |
| Insertion | Chr5 | 21036906 | 21036906 | 1 | |
| Insertion | Chr5 | 21280846 | 21280847 | 2 | |
| Insertion | Chr5 | 23445280 | 23445281 | 2 | |
| Insertion | Chr6 | 2340869 | 2340869 | 1 | |
| Insertion | Chr7 | 27544679 | 27544679 | 1 | |
| Insertion | Chr8 | 774395 | 774400 | 6 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 775449 | 1362370 | LOC_Os12g03470 |
| Inversion | Chr8 | 18818928 | 19219172 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 1207606 | Chr4 | 14957132 | 2 |
| Translocation | Chr11 | 1986111 | Chr1 | 40935919 | |
| Translocation | Chr11 | 1986283 | Chr1 | 40935911 | |
| Translocation | Chr12 | 4372113 | Chr6 | 21999589 | LOC_Os06g37250 |
| Translocation | Chr11 | 13157035 | Chr3 | 6684690 | |
| Translocation | Chr11 | 13157045 | Chr3 | 6689547 | LOC_Os03g12600 |
| Translocation | Chr11 | 21327648 | Chr3 | 25354092 | |
| Translocation | Chr11 | 27466140 | Chr5 | 11752869 |
