Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1630-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1630-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 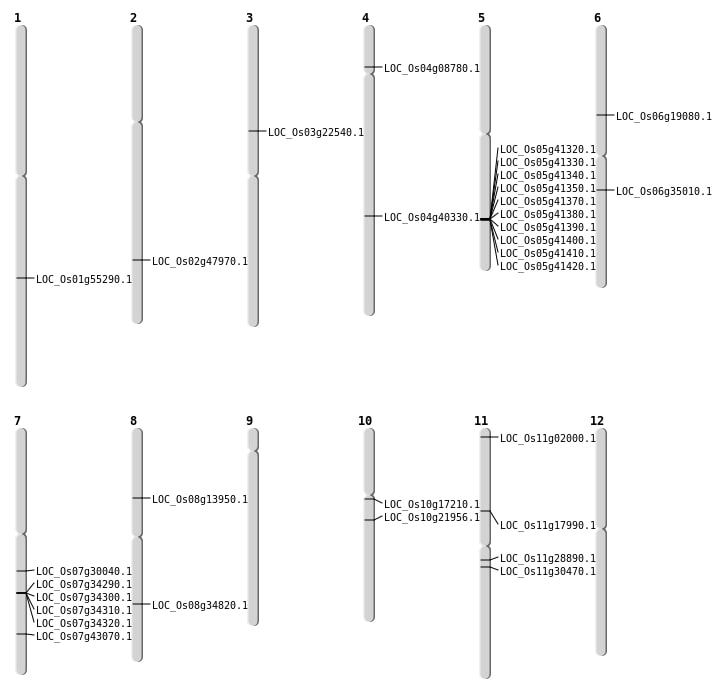
|
| Variant Information |
|---|
Single base substitutions: 53
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21792679 | G-A | Homo | INTERGENIC | |
| SBS | Chr10 | 16604583 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 20697641 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 10111933 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g17990 |
| SBS | Chr11 | 14100823 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 17716746 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30470 |
| SBS | Chr11 | 20218222 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 20783228 | T-A | Homo | INTERGENIC | |
| SBS | Chr11 | 9514556 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 17435778 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 18712657 | G-T | HET | INTERGENIC | |
| SBS | Chr12 | 20406476 | C-A | HET | INTERGENIC | |
| SBS | Chr12 | 8264907 | T-C | HET | INTERGENIC | |
| SBS | Chr2 | 11510302 | T-G | Homo | INTRON | |
| SBS | Chr2 | 27517627 | G-C | Homo | INTERGENIC | |
| SBS | Chr2 | 32212683 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 14005514 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 19830286 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 26924224 | C-T | HET | INTRON | |
| SBS | Chr3 | 470091 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 5117584 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 13888214 | G-A | Homo | INTERGENIC | |
| SBS | Chr4 | 16399900 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 21810413 | T-A | Homo | INTERGENIC | |
| SBS | Chr4 | 23979097 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g40330 |
| SBS | Chr5 | 10617848 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 18819226 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 20860796 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 24059929 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 24059933 | A-T | Homo | INTERGENIC | |
| SBS | Chr5 | 26290121 | A-C | Homo | INTRON | |
| SBS | Chr5 | 27404796 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 27633305 | C-T | HET | INTRON | |
| SBS | Chr5 | 6770482 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 20921471 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 7251190 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 12791736 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 17248992 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 18880290 | G-T | HET | INTERGENIC | |
| SBS | Chr7 | 18880291 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 18880296 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 26705339 | A-C | HET | INTERGENIC | |
| SBS | Chr7 | 27439607 | T-G | HET | INTERGENIC | |
| SBS | Chr7 | 9882076 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 14412828 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 16216144 | T-C | HET | INTERGENIC | |
| SBS | Chr8 | 16244820 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 24169735 | G-A | HET | INTERGENIC | |
| SBS | Chr8 | 6481005 | G-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr8 | 7618304 | C-T | HET | INTRON | |
| SBS | Chr8 | 8359394 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g13950 |
| SBS | Chr9 | 3785355 | G-T | HET | INTERGENIC | |
| SBS | Chr9 | 4165307 | C-A | HET | INTERGENIC |
Deletions: 43
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 533343 | 533351 | 8 | LOC_Os11g02000 |
| Deletion | Chr10 | 862650 | 862651 | 1 | |
| Deletion | Chr10 | 999850 | 999851 | 1 | |
| Deletion | Chr2 | 3743240 | 3743241 | 1 | |
| Deletion | Chr2 | 3869584 | 3869596 | 12 | |
| Deletion | Chr2 | 3980479 | 3980483 | 4 | |
| Deletion | Chr4 | 4789224 | 4789239 | 15 | LOC_Os04g08780 |
| Deletion | Chr5 | 5028003 | 5028024 | 21 | |
| Deletion | Chr3 | 6712562 | 6712568 | 6 | |
| Deletion | Chr5 | 6916972 | 6916973 | 1 | |
| Deletion | Chr6 | 7046530 | 7046538 | 8 | |
| Deletion | Chr10 | 8496727 | 8496733 | 6 | |
| Deletion | Chr10 | 8643797 | 8651329 | 7532 | LOC_Os10g17210 |
| Deletion | Chr1 | 8796797 | 8796799 | 2 | |
| Deletion | Chr8 | 10571500 | 10571507 | 7 | |
| Deletion | Chr11 | 11794048 | 11794056 | 8 | |
| Deletion | Chr11 | 12274591 | 12274708 | 117 | |
| Deletion | Chr3 | 12952482 | 12952485 | 3 | LOC_Os03g22540 |
| Deletion | Chr1 | 13746962 | 13746964 | 2 | |
| Deletion | Chr10 | 15021465 | 15021467 | 2 | |
| Deletion | Chr1 | 15073296 | 15073304 | 8 | |
| Deletion | Chr12 | 15551672 | 15551678 | 6 | |
| Deletion | Chr12 | 16105803 | 16105906 | 103 | |
| Deletion | Chr5 | 16301721 | 16301724 | 3 | |
| Deletion | Chr7 | 16329665 | 16329666 | 1 | |
| Deletion | Chr11 | 16720900 | 16720901 | 1 | LOC_Os11g28890 |
| Deletion | Chr7 | 17706686 | 17706693 | 7 | LOC_Os07g30040 |
| Deletion | Chr11 | 18422529 | 18422543 | 14 | |
| Deletion | Chr3 | 19322985 | 19322996 | 11 | |
| Deletion | Chr2 | 20313537 | 20313538 | 1 | |
| Deletion | Chr6 | 20359975 | 20361275 | 1300 | LOC_Os06g35010 |
| Deletion | Chr7 | 20530001 | 20570000 | 40000 | 4 |
| Deletion | Chr7 | 21181788 | 21181789 | 1 | |
| Deletion | Chr9 | 21640545 | 21640566 | 21 | |
| Deletion | Chr8 | 21906458 | 21906480 | 22 | LOC_Os08g34820 |
| Deletion | Chr7 | 23972585 | 23972586 | 1 | |
| Deletion | Chr5 | 24202001 | 24255000 | 53000 | 10 |
| Deletion | Chr7 | 25804528 | 25804529 | 1 | LOC_Os07g43070 |
| Deletion | Chr2 | 29344603 | 29344612 | 9 | LOC_Os02g47970 |
| Deletion | Chr1 | 30421641 | 30421654 | 13 | |
| Deletion | Chr1 | 31828417 | 31828420 | 3 | LOC_Os01g55290 |
| Deletion | Chr3 | 33609841 | 33609844 | 3 | |
| Deletion | Chr3 | 34182184 | 34182189 | 5 |
Insertions: 17
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 11338822 | Chr6 | 10850162 | 2 |
| Translocation | Chr6 | 12594675 | Chr1 | 32531298 |
