Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1631-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1631-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 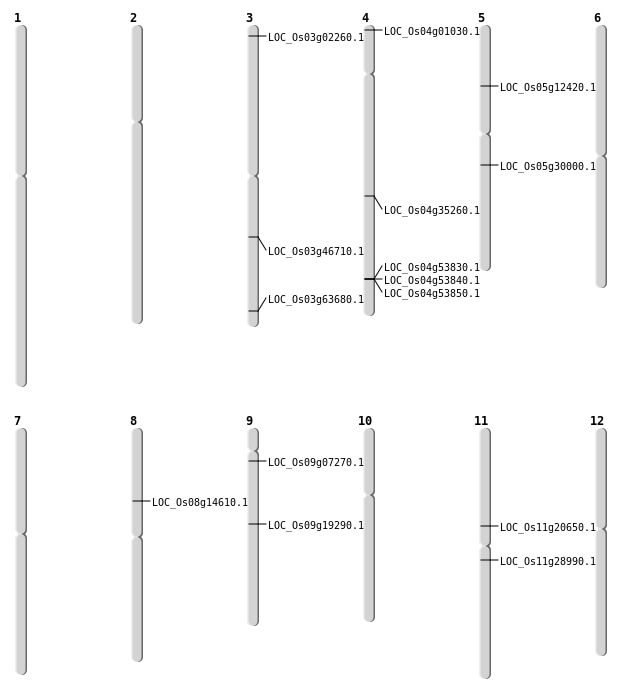
|
| Variant Information |
|---|
Single base substitutions: 25
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr11 | 3212600 | C-A | Homo | INTERGENIC | |
| SBS | Chr11 | 8688934 | C-G | HET | INTERGENIC | |
| SBS | Chr12 | 20041781 | A-T | HET | INTERGENIC | |
| SBS | Chr2 | 29517288 | C-A | Homo | INTRON | |
| SBS | Chr2 | 5504339 | A-T | Homo | UTR_3_PRIME | |
| SBS | Chr3 | 22135528 | C-A | Homo | INTERGENIC | |
| SBS | Chr3 | 24667470 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 26435826 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g46710 |
| SBS | Chr3 | 767187 | G-A | HET | STOP_GAINED | LOC_Os03g02260 |
| SBS | Chr4 | 21430582 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g35260 |
| SBS | Chr4 | 27836982 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr4 | 30175 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g01030 |
| SBS | Chr4 | 6122695 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6654166 | A-G | HET | INTERGENIC | |
| SBS | Chr4 | 9624138 | T-A | HET | INTERGENIC | |
| SBS | Chr5 | 15957230 | C-T | HET | INTERGENIC | |
| SBS | Chr5 | 25614335 | C-T | Homo | INTERGENIC | |
| SBS | Chr5 | 25614336 | G-T | Homo | INTERGENIC | |
| SBS | Chr5 | 7142261 | C-T | HET | STOP_GAINED | LOC_Os05g12420 |
| SBS | Chr6 | 11230724 | T-C | Homo | INTRON | |
| SBS | Chr7 | 20473670 | A-G | Homo | INTERGENIC | |
| SBS | Chr8 | 5916606 | G-C | Homo | INTERGENIC | |
| SBS | Chr9 | 11545182 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os09g19290 |
| SBS | Chr9 | 18675014 | C-T | Homo | INTERGENIC | |
| SBS | Chr9 | 3564110 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07270 |
Deletions: 30
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr5 | 2128896 | 2128897 | 1 | |
| Deletion | Chr8 | 4371349 | 4371350 | 1 | |
| Deletion | Chr10 | 4997389 | 4997398 | 9 | |
| Deletion | Chr4 | 6099069 | 6099072 | 3 | |
| Deletion | Chr5 | 6157368 | 6157369 | 1 | |
| Deletion | Chr10 | 6839709 | 6839724 | 15 | |
| Deletion | Chr8 | 9039646 | 9039661 | 15 | |
| Deletion | Chr2 | 10618494 | 10618496 | 2 | |
| Deletion | Chr1 | 10710589 | 10710590 | 1 | |
| Deletion | Chr6 | 11691561 | 11691569 | 8 | |
| Deletion | Chr7 | 12783048 | 12783050 | 2 | |
| Deletion | Chr5 | 12840078 | 12840079 | 1 | |
| Deletion | Chr1 | 13309878 | 13309879 | 1 | |
| Deletion | Chr4 | 13792669 | 13792675 | 6 | |
| Deletion | Chr3 | 14488408 | 14488409 | 1 | |
| Deletion | Chr8 | 15928590 | 15928940 | 350 | |
| Deletion | Chr12 | 16861037 | 16861038 | 1 | |
| Deletion | Chr5 | 17348988 | 17354772 | 5784 | LOC_Os05g30000 |
| Deletion | Chr12 | 19415582 | 19415651 | 69 | |
| Deletion | Chr8 | 20082429 | 20082436 | 7 | |
| Deletion | Chr2 | 20707437 | 20707438 | 1 | |
| Deletion | Chr11 | 22320166 | 22320168 | 2 | |
| Deletion | Chr1 | 22615700 | 22615703 | 3 | |
| Deletion | Chr6 | 24483020 | 24483021 | 1 | |
| Deletion | Chr2 | 26716943 | 26717014 | 71 | |
| Deletion | Chr3 | 30697728 | 30697730 | 2 | |
| Deletion | Chr4 | 32078764 | 32085857 | 7093 | 3 |
| Deletion | Chr3 | 33424761 | 33424763 | 2 | |
| Deletion | Chr3 | 35970240 | 35970252 | 12 | LOC_Os03g63680 |
| Deletion | Chr1 | 36152813 | 36152826 | 13 |
Insertions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 25245398 | 25245399 | 2 | |
| Insertion | Chr10 | 757685 | 757685 | 1 | |
| Insertion | Chr11 | 11970285 | 11970285 | 1 | LOC_Os11g20650 |
| Insertion | Chr11 | 16779721 | 16779721 | 1 | LOC_Os11g28990 |
| Insertion | Chr11 | 7305659 | 7305660 | 2 | |
| Insertion | Chr5 | 10166805 | 10166806 | 2 | |
| Insertion | Chr7 | 3839375 | 3839375 | 1 | |
| Insertion | Chr8 | 23428968 | 23428968 | 1 | |
| Insertion | Chr9 | 4781890 | 4781895 | 6 |
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 8790761 | 11775504 | LOC_Os08g14610 |
| Inversion | Chr8 | 8790777 | 10588636 | LOC_Os08g14610 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 8214649 | Chr5 | 29794244 | |
| Translocation | Chr12 | 8494119 | Chr4 | 17935005 | LOC_Os04g30040 |
