Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1633-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1633-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 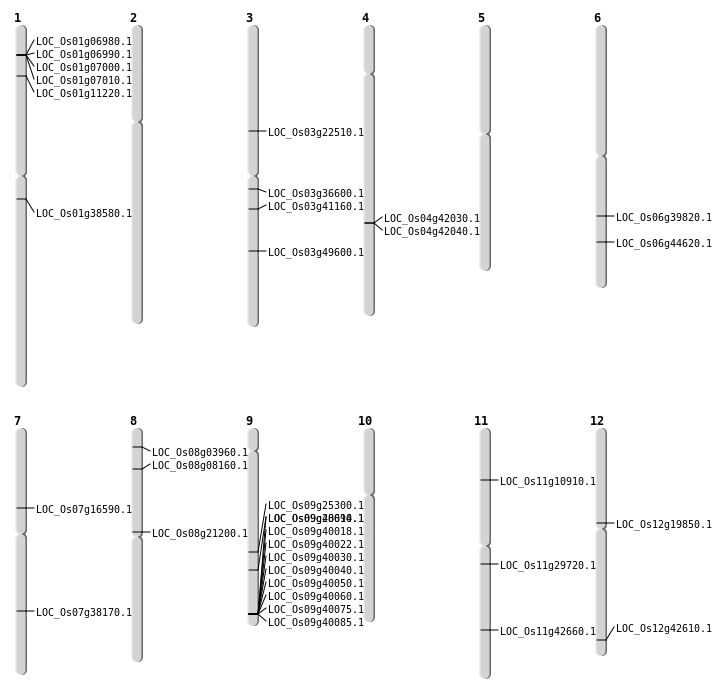
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16290143 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 17840184 | T-G | HET | INTERGENIC | |
| SBS | Chr1 | 22458832 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 5070091 | A-G | HET | INTERGENIC | |
| SBS | Chr1 | 6748941 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr10 | 11961727 | G-A | HET | INTERGENIC | |
| SBS | Chr10 | 11961729 | G-T | HET | INTERGENIC | |
| SBS | Chr11 | 13077015 | A-C | HET | INTERGENIC | |
| SBS | Chr11 | 17237260 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g29720 |
| SBS | Chr11 | 25696881 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g42660 |
| SBS | Chr11 | 5959404 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 977852 | C-T | HET | INTRON | |
| SBS | Chr12 | 11575156 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19850 |
| SBS | Chr12 | 1431062 | G-T | HET | INTRON | |
| SBS | Chr12 | 24302524 | C-A | HET | INTERGENIC | |
| SBS | Chr2 | 15354544 | G-A | HET | INTERGENIC | |
| SBS | Chr2 | 29735826 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 19590162 | C-T | HET | INTRON | |
| SBS | Chr3 | 20308688 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 20723114 | T-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr3 | 22884860 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g41160 |
| SBS | Chr3 | 5676925 | G-A | HET | INTERGENIC | |
| SBS | Chr3 | 8823254 | A-T | Homo | INTRON | |
| SBS | Chr4 | 23144813 | C-A | Homo | INTERGENIC | |
| SBS | Chr4 | 28748525 | G-A | HET | INTERGENIC | |
| SBS | Chr5 | 22168340 | C-G | HET | INTERGENIC | |
| SBS | Chr5 | 26747466 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 16563951 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 17219009 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 17219010 | T-A | HET | INTERGENIC | |
| SBS | Chr6 | 17219011 | G-A | HET | INTERGENIC | |
| SBS | Chr6 | 17559798 | A-G | HET | INTRON | |
| SBS | Chr6 | 23659070 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g39820 |
| SBS | Chr6 | 26874575 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 10454325 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 14768018 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 15751722 | G-T | HET | INTRON | |
| SBS | Chr7 | 17654083 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 20349655 | A-T | HET | INTERGENIC | |
| SBS | Chr7 | 22890902 | T-A | HET | UTR_5_PRIME | |
| SBS | Chr8 | 12656961 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21200 |
| SBS | Chr8 | 25187103 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 6054575 | G-C | Homo | INTERGENIC | |
| SBS | Chr9 | 14224490 | G-C | Homo | INTERGENIC | |
| SBS | Chr9 | 1903509 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 19722499 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 2978993 | A-C | HET | INTERGENIC | |
| SBS | Chr9 | 3451098 | A-G | HET | INTRON | |
| SBS | Chr9 | 8432713 | A-T | HET | INTRON | |
| SBS | Chr9 | 8432714 | G-T | HET | INTRON | |
| SBS | Chr9 | 9207900 | A-T | HET | SPLICE_SITE_REGION |
Deletions: 48
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 39683 | 39687 | 4 | |
| Deletion | Chr9 | 2981449 | 2981464 | 15 | |
| Deletion | Chr1 | 3294800 | 3303681 | 8881 | 4 |
| Deletion | Chr2 | 4170171 | 4170172 | 1 | |
| Deletion | Chr2 | 4238242 | 4238250 | 8 | |
| Deletion | Chr8 | 4345404 | 4345423 | 19 | |
| Deletion | Chr8 | 4594516 | 4594518 | 2 | |
| Deletion | Chr8 | 6111034 | 6111044 | 10 | |
| Deletion | Chr7 | 6388782 | 6388787 | 5 | |
| Deletion | Chr1 | 7318110 | 7318111 | 1 | |
| Deletion | Chr6 | 7785039 | 7785040 | 1 | |
| Deletion | Chr10 | 8096412 | 8096414 | 2 | |
| Deletion | Chr2 | 8288386 | 8288387 | 1 | |
| Deletion | Chr6 | 8716540 | 8716548 | 8 | |
| Deletion | Chr7 | 9733911 | 9733916 | 5 | LOC_Os07g16590 |
| Deletion | Chr7 | 10347316 | 10347322 | 6 | |
| Deletion | Chr11 | 11517501 | 11517512 | 11 | |
| Deletion | Chr10 | 12300605 | 12300606 | 1 | |
| Deletion | Chr1 | 12755267 | 12755268 | 1 | |
| Deletion | Chr10 | 13159516 | 13159517 | 1 | |
| Deletion | Chr9 | 13724000 | 13724004 | 4 | |
| Deletion | Chr10 | 14050798 | 14050799 | 1 | |
| Deletion | Chr7 | 14486480 | 14486482 | 2 | |
| Deletion | Chr9 | 15145204 | 15145293 | 89 | LOC_Os09g25300 |
| Deletion | Chr7 | 15837631 | 15837641 | 10 | |
| Deletion | Chr8 | 17273465 | 17273466 | 1 | |
| Deletion | Chr7 | 17414800 | 17414801 | 1 | |
| Deletion | Chr9 | 17449376 | 17449378 | 2 | LOC_Os09g28690 |
| Deletion | Chr6 | 17702748 | 17702811 | 63 | |
| Deletion | Chr10 | 19424231 | 19424237 | 6 | |
| Deletion | Chr6 | 20195736 | 20195745 | 9 | |
| Deletion | Chr3 | 20294886 | 20294903 | 17 | LOC_Os03g36600 |
| Deletion | Chr2 | 20302601 | 20304380 | 1779 | |
| Deletion | Chr11 | 20545599 | 20545600 | 1 | |
| Deletion | Chr10 | 20569918 | 20569940 | 22 | |
| Deletion | Chr3 | 21293852 | 21293853 | 1 | |
| Deletion | Chr1 | 21667735 | 21667744 | 9 | LOC_Os01g38580 |
| Deletion | Chr8 | 22252494 | 22252505 | 11 | |
| Deletion | Chr9 | 22962001 | 23013000 | 51000 | 9 |
| Deletion | Chr4 | 24893837 | 24903051 | 9214 | 2 |
| Deletion | Chr4 | 25809769 | 25809772 | 3 | |
| Deletion | Chr2 | 25874843 | 25874850 | 7 | |
| Deletion | Chr12 | 26477776 | 26477778 | 2 | LOC_Os12g42610 |
| Deletion | Chr7 | 26767343 | 26767344 | 1 | |
| Deletion | Chr6 | 26945523 | 26945539 | 16 | LOC_Os06g44620 |
| Deletion | Chr2 | 27196339 | 27196348 | 9 | |
| Deletion | Chr3 | 28241822 | 28241831 | 9 | LOC_Os03g49600 |
| Deletion | Chr1 | 30509188 | 30509198 | 10 |
Insertions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 6021022 | 6021025 | 4 | LOC_Os11g10910 |
| Insertion | Chr12 | 16737096 | 16737096 | 1 | |
| Insertion | Chr3 | 12930154 | 12930171 | 18 | LOC_Os03g22510 |
| Insertion | Chr4 | 19542729 | 19542729 | 1 | |
| Insertion | Chr5 | 12042218 | 12042218 | 1 | |
| Insertion | Chr5 | 7539901 | 7539901 | 1 | |
| Insertion | Chr5 | 7814999 | 7815000 | 2 | |
| Insertion | Chr6 | 10581862 | 10581863 | 2 | |
| Insertion | Chr7 | 11319117 | 11319118 | 2 | |
| Insertion | Chr7 | 201046 | 201047 | 2 | |
| Insertion | Chr8 | 10375796 | 10375796 | 1 | |
| Insertion | Chr8 | 27428570 | 27428573 | 4 | |
| Insertion | Chr8 | 9284999 | 9285004 | 6 | |
| Insertion | Chr9 | 18046414 | 18046415 | 2 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 762339 | 1889266 | LOC_Os08g03960 |
| Inversion | Chr1 | 5822535 | 6005259 | LOC_Os01g11220 |
| Inversion | Chr1 | 5890477 | 6005257 | LOC_Os01g11220 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 4646981 | Chr7 | 22890621 | 2 |
| Translocation | Chr8 | 4647481 | Chr7 | 22890615 | 2 |
| Translocation | Chr8 | 12681547 | Chr4 | 3704776 |
