Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1638-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1638-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 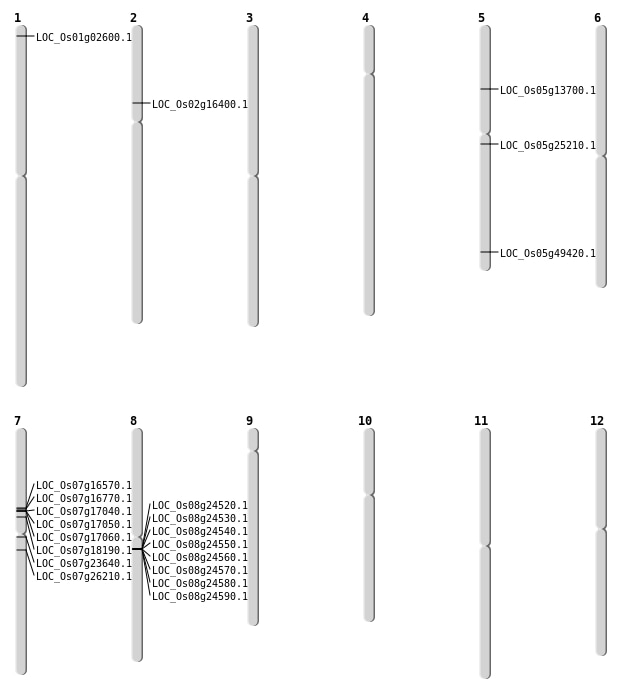
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 27226548 | C-T | HET | UTR_3_PRIME | |
| SBS | Chr1 | 40380146 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 866679 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g02600 |
| SBS | Chr10 | 13397231 | C-A | HET | INTRON | |
| SBS | Chr11 | 20766034 | G-A | Homo | INTRON | |
| SBS | Chr11 | 378053 | G-A | Homo | INTERGENIC | |
| SBS | Chr12 | 12387847 | T-G | HET | INTRON | |
| SBS | Chr12 | 19386657 | A-G | HET | SYNONYMOUS_CODING | |
| SBS | Chr12 | 20427582 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 24527560 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr12 | 24726149 | G-T | Homo | INTERGENIC | |
| SBS | Chr12 | 27372356 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 27589543 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 9349022 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16400 |
| SBS | Chr3 | 2860921 | G-A | Homo | INTERGENIC | |
| SBS | Chr3 | 35352370 | A-C | HET | INTRON | |
| SBS | Chr4 | 2214545 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 27331286 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 27920078 | A-T | HET | INTRON | |
| SBS | Chr4 | 28891450 | T-C | HET | INTERGENIC | |
| SBS | Chr5 | 15923167 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 17227409 | A-T | Homo | INTERGENIC | |
| SBS | Chr5 | 28338780 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g49420 |
| SBS | Chr5 | 28484830 | G-T | HET | INTERGENIC | |
| SBS | Chr5 | 7286847 | A-G | HET | INTERGENIC | |
| SBS | Chr6 | 16398731 | G-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr6 | 16398732 | C-T | Homo | SYNONYMOUS_CODING | |
| SBS | Chr6 | 20533964 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 10787587 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18190 |
| SBS | Chr7 | 11838885 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 13843551 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 17654062 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 5504914 | G-A | Homo | INTRON | |
| SBS | Chr8 | 13573487 | C-T | HET | INTRON | |
| SBS | Chr8 | 16175887 | A-G | HET | INTERGENIC | |
| SBS | Chr8 | 8783 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 1297502 | T-C | HET | INTERGENIC | |
| SBS | Chr9 | 17071614 | T-C | Homo | INTERGENIC |
Deletions: 26
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 1491105 | 1491106 | 1 | |
| Deletion | Chr3 | 2457601 | 2457603 | 2 | |
| Deletion | Chr4 | 3477164 | 3477165 | 1 | |
| Deletion | Chr2 | 5199535 | 5199536 | 1 | |
| Deletion | Chr5 | 7590379 | 7590382 | 3 | |
| Deletion | Chr4 | 7962920 | 7962921 | 1 | |
| Deletion | Chr7 | 8582113 | 8582149 | 36 | |
| Deletion | Chr11 | 9566368 | 9566369 | 1 | |
| Deletion | Chr7 | 9719616 | 9720401 | 785 | LOC_Os07g16570 |
| Deletion | Chr7 | 9843119 | 9843121 | 2 | LOC_Os07g16770 |
| Deletion | Chr7 | 10033680 | 10041997 | 8317 | 3 |
| Deletion | Chr6 | 10230492 | 10230512 | 20 | |
| Deletion | Chr7 | 11772048 | 11772051 | 3 | |
| Deletion | Chr7 | 13354547 | 13354548 | 1 | LOC_Os07g23640 |
| Deletion | Chr7 | 14569546 | 14569551 | 5 | |
| Deletion | Chr5 | 14614374 | 14614375 | 1 | LOC_Os05g25210 |
| Deletion | Chr8 | 14809001 | 14875000 | 66000 | 8 |
| Deletion | Chr7 | 15075300 | 15075320 | 20 | LOC_Os07g26210 |
| Deletion | Chr2 | 19619537 | 19619551 | 14 | |
| Deletion | Chr6 | 20779636 | 20779638 | 2 | |
| Deletion | Chr2 | 22376486 | 22376490 | 4 | |
| Deletion | Chr12 | 22841356 | 22841358 | 2 | |
| Deletion | Chr6 | 24845788 | 24845789 | 1 | |
| Deletion | Chr3 | 31757262 | 31757263 | 1 | |
| Deletion | Chr3 | 31888018 | 31888021 | 3 | |
| Deletion | Chr1 | 41028458 | 41028462 | 4 |
Insertions: 12
Inversions: 4
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 7147866 | 7592420 | LOC_Os05g13700 |
| Inversion | Chr3 | 25920385 | 27057022 | |
| Inversion | Chr3 | 26638153 | 26812166 | |
| Inversion | Chr3 | 26638222 | 26812176 |
No Translocation
