Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1646-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1646-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 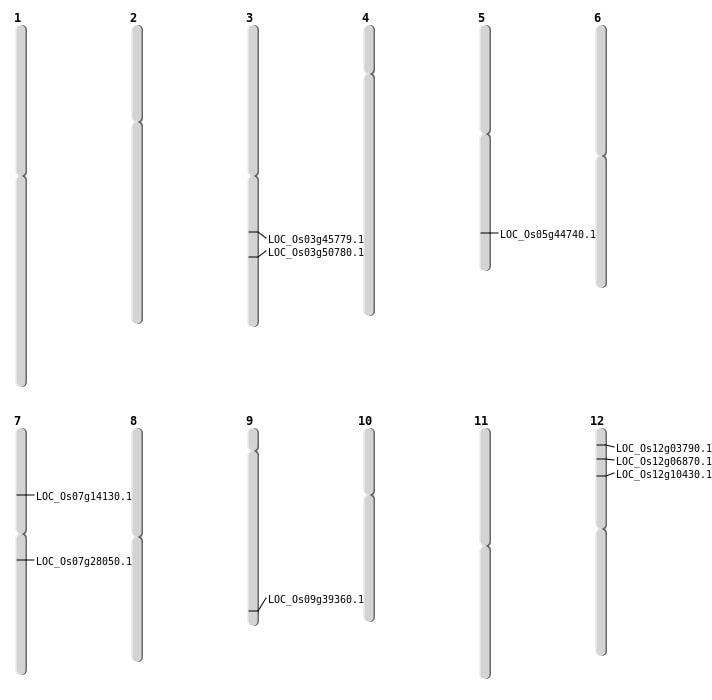
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12257844 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 29097901 | A-G | Homo | INTERGENIC | |
| SBS | Chr10 | 20074557 | A-T | Homo | INTERGENIC | |
| SBS | Chr11 | 18913826 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 24841923 | C-G | HET | INTRON | |
| SBS | Chr12 | 8752602 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr2 | 14667059 | C-T | HET | INTRON | |
| SBS | Chr2 | 17877305 | T-G | HET | INTERGENIC | |
| SBS | Chr3 | 14053017 | C-T | HET | INTERGENIC | |
| SBS | Chr3 | 14199040 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 10751234 | C-T | HET | INTERGENIC | |
| SBS | Chr4 | 11128616 | G-C | HET | INTERGENIC | |
| SBS | Chr4 | 14467616 | G-T | HET | INTERGENIC | |
| SBS | Chr4 | 3002231 | G-C | HET | INTRON | |
| SBS | Chr4 | 32918719 | T-A | HET | INTRON | |
| SBS | Chr5 | 20495472 | C-A | Homo | INTERGENIC | |
| SBS | Chr5 | 26010938 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g44740 |
| SBS | Chr7 | 10472637 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 22087409 | C-A | HET | INTERGENIC | |
| SBS | Chr8 | 2830510 | G-A | Homo | INTERGENIC | |
| SBS | Chr8 | 2830511 | T-A | Homo | INTERGENIC | |
| SBS | Chr9 | 20519470 | C-T | HET | SPLICE_SITE_REGION | |
| SBS | Chr9 | 2061682 | G-A | HET | INTERGENIC | |
| SBS | Chr9 | 22635926 | C-T | HET | START_GAINED | LOC_Os09g39360 |
| SBS | Chr9 | 2941905 | C-G | HET | INTERGENIC | |
| SBS | Chr9 | 3771714 | G-A | HET | INTERGENIC |
Deletions: 38
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1943491 | 1943492 | 1 | |
| Deletion | Chr3 | 2595408 | 2595427 | 19 | |
| Deletion | Chr9 | 4559945 | 4559948 | 3 | |
| Deletion | Chr12 | 4560232 | 4560245 | 13 | |
| Deletion | Chr9 | 5166966 | 5167019 | 53 | |
| Deletion | Chr12 | 5525432 | 5525442 | 10 | LOC_Os12g10430 |
| Deletion | Chr7 | 8064178 | 8064182 | 4 | LOC_Os07g14130 |
| Deletion | Chr2 | 9534895 | 9534917 | 22 | |
| Deletion | Chr12 | 10958531 | 10958532 | 1 | |
| Deletion | Chr3 | 11157832 | 11157846 | 14 | |
| Deletion | Chr11 | 11283781 | 11283782 | 1 | |
| Deletion | Chr5 | 11621156 | 11621158 | 2 | |
| Deletion | Chr4 | 11760723 | 11760726 | 3 | |
| Deletion | Chr8 | 11839660 | 11839676 | 16 | |
| Deletion | Chr2 | 12266196 | 12266204 | 8 | |
| Deletion | Chr4 | 12527012 | 12527020 | 8 | |
| Deletion | Chr5 | 12558658 | 12558678 | 20 | |
| Deletion | Chr11 | 13137881 | 13137893 | 12 | |
| Deletion | Chr7 | 15381541 | 15381542 | 1 | |
| Deletion | Chr7 | 15643340 | 15643353 | 13 | |
| Deletion | Chr2 | 16731412 | 16731413 | 1 | |
| Deletion | Chr3 | 16747972 | 16747980 | 8 | |
| Deletion | Chr4 | 17092637 | 17092641 | 4 | |
| Deletion | Chr7 | 17219209 | 17220876 | 1667 | |
| Deletion | Chr4 | 17582026 | 17582027 | 1 | |
| Deletion | Chr9 | 18543215 | 18543226 | 11 | |
| Deletion | Chr5 | 20495462 | 20495465 | 3 | |
| Deletion | Chr4 | 20618522 | 20618529 | 7 | |
| Deletion | Chr8 | 21813709 | 21813710 | 1 | |
| Deletion | Chr5 | 21899887 | 21899889 | 2 | |
| Deletion | Chr9 | 22711497 | 22711498 | 1 | |
| Deletion | Chr12 | 23632025 | 23632027 | 2 | |
| Deletion | Chr3 | 25859666 | 25859667 | 1 | LOC_Os03g45779 |
| Deletion | Chr1 | 26704539 | 26704540 | 1 | |
| Deletion | Chr1 | 34513856 | 34513868 | 12 | |
| Deletion | Chr1 | 38075994 | 38075998 | 4 | |
| Deletion | Chr1 | 38904274 | 38904276 | 2 | |
| Deletion | Chr1 | 42289269 | 42289271 | 2 |
Insertions: 12
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 1542159 | 3339422 | 2 |
| Inversion | Chr12 | 1542173 | 3339411 | 2 |
| Inversion | Chr12 | 14631624 | 14853206 | |
| Inversion | Chr7 | 15575748 | 16361517 | LOC_Os07g28050 |
| Inversion | Chr7 | 15579235 | 16361516 | LOC_Os07g28050 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 4078638 | Chr3 | 28990577 | LOC_Os03g50780 |
| Translocation | Chr9 | 4079465 | Chr3 | 28990533 | LOC_Os03g50780 |
| Translocation | Chr12 | 9725470 | Chr9 | 13612940 | |
| Translocation | Chr6 | 15747401 | Chr4 | 19596843 |
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr7 | 15579233 | 17220862 | 1641629 |
