Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1651-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1651-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 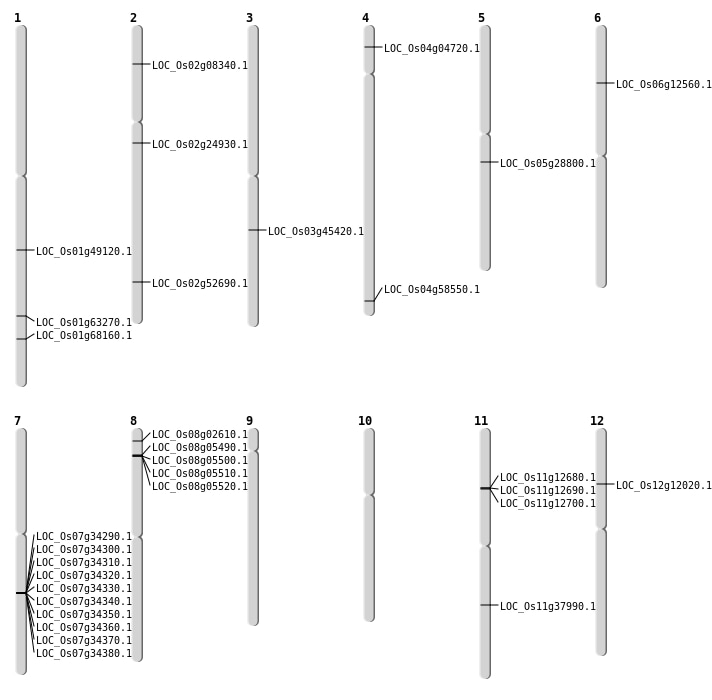
|
| Variant Information |
|---|
Single base substitutions: 28
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 21336361 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 28237059 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g49120 |
| SBS | Chr1 | 29177926 | G-A | HET | INTERGENIC | |
| SBS | Chr1 | 36675508 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g63270 |
| SBS | Chr10 | 17651306 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 12972810 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 147115 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 3944508 | G-A | HET | INTERGENIC | |
| SBS | Chr12 | 6590605 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g12020 |
| SBS | Chr2 | 32030635 | C-T | Homo | INTRON | |
| SBS | Chr2 | 3988849 | A-G | HET | INTERGENIC | |
| SBS | Chr2 | 4429006 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g08340 |
| SBS | Chr2 | 4429007 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g08340 |
| SBS | Chr3 | 28339182 | G-T | HET | INTERGENIC | |
| SBS | Chr3 | 295595 | T-C | Homo | UTR_3_PRIME | |
| SBS | Chr4 | 18946047 | T-C | HET | INTERGENIC | |
| SBS | Chr4 | 2269511 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g04720 |
| SBS | Chr4 | 34600717 | A-T | HET | INTERGENIC | |
| SBS | Chr4 | 34811673 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g58550 |
| SBS | Chr5 | 16885890 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g28800 |
| SBS | Chr7 | 26677878 | A-G | HET | INTRON | |
| SBS | Chr7 | 27740125 | C-A | HET | INTERGENIC | |
| SBS | Chr7 | 5651941 | C-T | Homo | INTERGENIC | |
| SBS | Chr7 | 6357021 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 15652099 | G-T | HET | INTERGENIC | |
| SBS | Chr8 | 18756954 | T-A | HET | INTERGENIC | |
| SBS | Chr8 | 7010340 | G-T | HET | INTRON | |
| SBS | Chr9 | 4543785 | G-T | HET | INTERGENIC |
Deletions: 34
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 1082053 | 1082054 | 1 | LOC_Os08g02610 |
| Deletion | Chr8 | 2924001 | 2960000 | 36000 | 4 |
| Deletion | Chr5 | 5374837 | 5374845 | 8 | |
| Deletion | Chr6 | 6809703 | 6809704 | 1 | LOC_Os06g12560 |
| Deletion | Chr11 | 7143001 | 7173000 | 30000 | 3 |
| Deletion | Chr4 | 9615299 | 9615304 | 5 | |
| Deletion | Chr12 | 10616980 | 10616987 | 7 | |
| Deletion | Chr9 | 11379641 | 11379650 | 9 | |
| Deletion | Chr7 | 12850502 | 12850538 | 36 | |
| Deletion | Chr5 | 13437957 | 13437960 | 3 | |
| Deletion | Chr10 | 14021197 | 14025222 | 4025 | |
| Deletion | Chr2 | 14453956 | 14453960 | 4 | LOC_Os02g24930 |
| Deletion | Chr10 | 15083802 | 15083824 | 22 | |
| Deletion | Chr4 | 15966211 | 15966255 | 44 | |
| Deletion | Chr5 | 16885296 | 16885383 | 87 | |
| Deletion | Chr5 | 16958920 | 16958932 | 12 | |
| Deletion | Chr3 | 17798861 | 17798877 | 16 | |
| Deletion | Chr12 | 18027836 | 18027841 | 5 | |
| Deletion | Chr12 | 18035212 | 18035217 | 5 | |
| Deletion | Chr9 | 19210546 | 19210553 | 7 | |
| Deletion | Chr2 | 19634333 | 19634334 | 1 | |
| Deletion | Chr6 | 19743780 | 19743785 | 5 | |
| Deletion | Chr7 | 20529001 | 20607000 | 78000 | 10 |
| Deletion | Chr1 | 21336356 | 21336357 | 1 | |
| Deletion | Chr1 | 22136040 | 22136049 | 9 | |
| Deletion | Chr11 | 22524136 | 22524903 | 767 | LOC_Os11g37990 |
| Deletion | Chr12 | 22856904 | 22856936 | 32 | |
| Deletion | Chr12 | 22941095 | 22941096 | 1 | |
| Deletion | Chr4 | 25606725 | 25606726 | 1 | |
| Deletion | Chr3 | 25638672 | 25638675 | 3 | LOC_Os03g45420 |
| Deletion | Chr8 | 25852694 | 25852695 | 1 | |
| Deletion | Chr8 | 26172295 | 26172307 | 12 | |
| Deletion | Chr2 | 32223953 | 32223954 | 1 | |
| Deletion | Chr2 | 32230211 | 32230224 | 13 | LOC_Os02g52690 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 24197549 | 24197549 | 1 | |
| Insertion | Chr2 | 26608218 | 26608221 | 4 | |
| Insertion | Chr2 | 29891598 | 29891598 | 1 | |
| Insertion | Chr5 | 15837807 | 15837812 | 6 |
No Inversion
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 3927134 | Chr1 | 39625738 | LOC_Os01g68160 |
| Translocation | Chr7 | 11836167 | Chr1 | 13837383 | |
| Translocation | Chr11 | 28214852 | Chr6 | 26628334 |
