Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1657-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1657-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 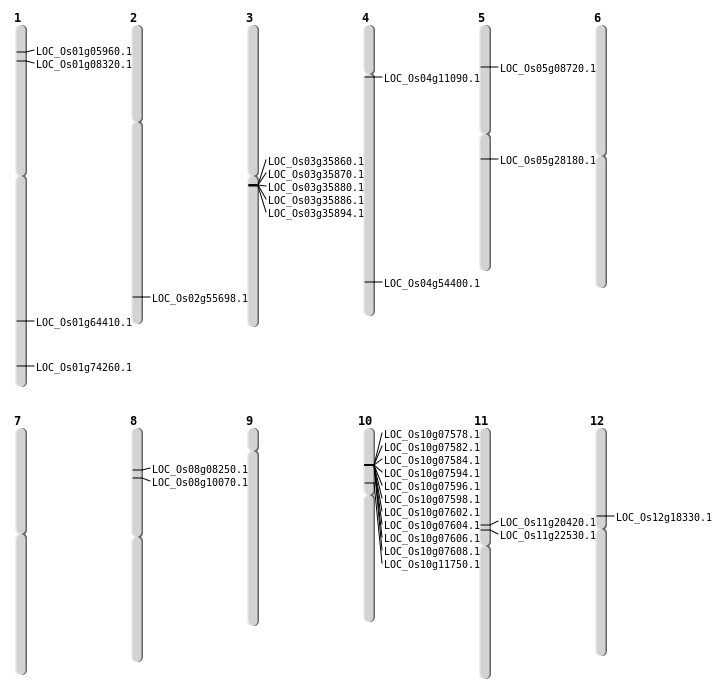
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 9574459 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 12945143 | C-A | HET | STOP_GAINED | LOC_Os11g22530 |
| SBS | Chr11 | 13929152 | C-T | HET | INTERGENIC | |
| SBS | Chr11 | 15126081 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 15342707 | T-C | HET | UTR_3_PRIME | |
| SBS | Chr12 | 15943162 | T-G | Homo | INTERGENIC | |
| SBS | Chr2 | 12827055 | G-A | Homo | INTRON | |
| SBS | Chr2 | 32398023 | A-T | Homo | INTERGENIC | |
| SBS | Chr3 | 17197108 | A-G | HET | INTERGENIC | |
| SBS | Chr3 | 20912052 | T-C | HET | INTERGENIC | |
| SBS | Chr3 | 29419488 | C-A | HET | INTERGENIC | |
| SBS | Chr3 | 5349378 | T-C | HET | INTRON | |
| SBS | Chr3 | 6806510 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr4 | 32368263 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g54400 |
| SBS | Chr5 | 12600206 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 13268651 | C-A | HET | INTERGENIC | |
| SBS | Chr6 | 15358021 | C-A | HET | INTRON | |
| SBS | Chr6 | 31094991 | A-G | Homo | INTERGENIC | |
| SBS | Chr7 | 21709894 | A-C | Homo | INTRON | |
| SBS | Chr7 | 6890549 | T-A | Homo | INTERGENIC | |
| SBS | Chr8 | 23046442 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 26937449 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 28181135 | C-A | HET | INTRON | |
| SBS | Chr8 | 8495175 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr9 | 15661595 | G-A | Homo | INTERGENIC | |
| SBS | Chr9 | 16361268 | C-A | Homo | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 1097680 | 1097688 | 8 | |
| Deletion | Chr11 | 2091754 | 2091763 | 9 | |
| Deletion | Chr10 | 4103001 | 4191000 | 88000 | 10 |
| Deletion | Chr8 | 4718348 | 4718359 | 11 | LOC_Os08g08250 |
| Deletion | Chr5 | 4780343 | 4780346 | 3 | LOC_Os05g08720 |
| Deletion | Chr8 | 5843854 | 5843859 | 5 | LOC_Os08g10070 |
| Deletion | Chr10 | 6226775 | 6226792 | 17 | |
| Deletion | Chr10 | 6510187 | 6510203 | 16 | LOC_Os10g11750 |
| Deletion | Chr2 | 6850813 | 6850836 | 23 | |
| Deletion | Chr8 | 6942174 | 6942175 | 1 | |
| Deletion | Chr4 | 7112959 | 7112961 | 2 | |
| Deletion | Chr12 | 7588384 | 7588545 | 161 | |
| Deletion | Chr6 | 8779996 | 8780008 | 12 | |
| Deletion | Chr12 | 9238951 | 9238955 | 4 | |
| Deletion | Chr4 | 9832108 | 9832112 | 4 | |
| Deletion | Chr6 | 10329237 | 10329238 | 1 | |
| Deletion | Chr12 | 10579475 | 10579477 | 2 | LOC_Os12g18330 |
| Deletion | Chr12 | 11764527 | 11764538 | 11 | |
| Deletion | Chr11 | 11816089 | 11816094 | 5 | LOC_Os11g20420 |
| Deletion | Chr6 | 12451735 | 12451747 | 12 | |
| Deletion | Chr3 | 13683886 | 13683887 | 1 | |
| Deletion | Chr11 | 14265367 | 14265368 | 1 | |
| Deletion | Chr2 | 14374643 | 14374646 | 3 | |
| Deletion | Chr7 | 19130278 | 19130300 | 22 | |
| Deletion | Chr9 | 19234642 | 19234655 | 13 | |
| Deletion | Chr3 | 19884001 | 19917000 | 33000 | 5 |
| Deletion | Chr2 | 19962404 | 19962405 | 1 | |
| Deletion | Chr3 | 20579976 | 20579988 | 12 | |
| Deletion | Chr12 | 20980969 | 20980971 | 2 | |
| Deletion | Chr4 | 21904768 | 21904776 | 8 | |
| Deletion | Chr7 | 25935551 | 25935554 | 3 | |
| Deletion | Chr3 | 26556621 | 26556635 | 14 | |
| Deletion | Chr5 | 29594295 | 29594299 | 4 | |
| Deletion | Chr2 | 34164920 | 34164925 | 5 | |
| Deletion | Chr1 | 37381723 | 37381724 | 1 | LOC_Os01g64410 |
| Deletion | Chr1 | 43031522 | 43031523 | 1 | LOC_Os01g74260 |
Insertions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19519264 | 19519269 | 6 | |
| Insertion | Chr10 | 11737241 | 11737254 | 14 | |
| Insertion | Chr10 | 22436907 | 22436907 | 1 | |
| Insertion | Chr11 | 6799259 | 6799259 | 1 | |
| Insertion | Chr12 | 19957888 | 19957893 | 6 | |
| Insertion | Chr2 | 34109073 | 34109073 | 1 | LOC_Os02g55698 |
| Insertion | Chr3 | 22117637 | 22117638 | 2 | |
| Insertion | Chr3 | 4882033 | 4882033 | 1 | |
| Insertion | Chr3 | 6525773 | 6525773 | 1 | |
| Insertion | Chr4 | 16789079 | 16789079 | 1 | |
| Insertion | Chr4 | 6033345 | 6033346 | 2 | LOC_Os04g11090 |
| Insertion | Chr4 | 8129447 | 8129447 | 1 | |
| Insertion | Chr4 | 8607098 | 8607138 | 41 | |
| Insertion | Chr5 | 9906759 | 9906759 | 1 | |
| Insertion | Chr8 | 18413261 | 18413262 | 2 | |
| Insertion | Chr9 | 19701113 | 19701113 | 1 |
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 2840059 | 4075206 | 2 |
| Inversion | Chr5 | 16189829 | 16483804 | LOC_Os05g28180 |
| Inversion | Chr5 | 16189843 | 16483818 | LOC_Os05g28180 |
No Translocation
