Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1662-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1662-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 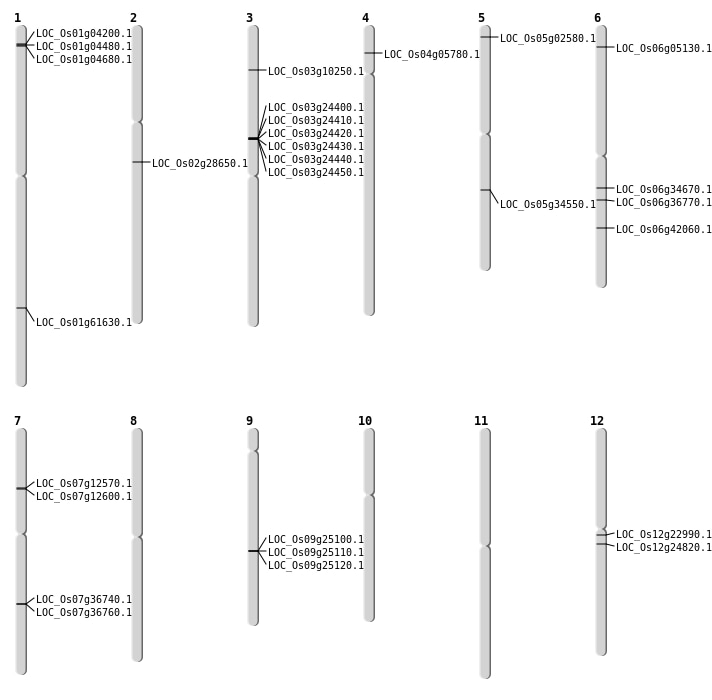
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 20175887 | C-T | Homo | INTRON | |
| SBS | Chr1 | 3936444 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 4239552 | T-C | HET | INTRON | |
| SBS | Chr1 | 4982892 | G-A | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 1208923 | A-T | HET | INTERGENIC | |
| SBS | Chr11 | 13476267 | C-T | HET | SYNONYMOUS_CODING | |
| SBS | Chr11 | 20718859 | A-G | HET | INTERGENIC | |
| SBS | Chr11 | 21282244 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 25517760 | C-T | HET | INTERGENIC | |
| SBS | Chr12 | 9590142 | G-A | HET | INTRON | |
| SBS | Chr2 | 17653375 | C-T | Homo | INTERGENIC | |
| SBS | Chr2 | 32207222 | C-T | Homo | INTERGENIC | |
| SBS | Chr3 | 5219487 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g10250 |
| SBS | Chr4 | 10843559 | C-A | HET | INTERGENIC | |
| SBS | Chr4 | 13564858 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 14512646 | A-G | HET | INTRON | |
| SBS | Chr4 | 16379547 | G-A | HET | INTERGENIC | |
| SBS | Chr4 | 6710518 | G-A | Homo | INTRON | |
| SBS | Chr5 | 10928313 | G-T | HET | INTRON | |
| SBS | Chr5 | 20175278 | C-A | HET | INTERGENIC | |
| SBS | Chr5 | 24693868 | G-A | Homo | SYNONYMOUS_CODING | |
| SBS | Chr6 | 18205781 | G-A | Homo | INTERGENIC | |
| SBS | Chr7 | 10940120 | T-A | HET | INTERGENIC | |
| SBS | Chr7 | 15181954 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 16622839 | A-T | HET | INTERGENIC | |
| SBS | Chr9 | 17085422 | C-T | HET | INTERGENIC |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 150418 | 150421 | 3 | |
| Deletion | Chr11 | 705182 | 705183 | 1 | |
| Deletion | Chr5 | 900512 | 900519 | 7 | LOC_Os05g02580 |
| Deletion | Chr5 | 2023480 | 2023481 | 1 | |
| Deletion | Chr6 | 3313607 | 3313610 | 3 | |
| Deletion | Chr12 | 3316240 | 3316250 | 10 | |
| Deletion | Chr9 | 3522002 | 3522016 | 14 | |
| Deletion | Chr9 | 3529312 | 3529314 | 2 | |
| Deletion | Chr12 | 4077631 | 4077637 | 6 | |
| Deletion | Chr6 | 4139289 | 4139290 | 1 | |
| Deletion | Chr4 | 4226850 | 4226853 | 3 | |
| Deletion | Chr11 | 5702559 | 5702560 | 1 | |
| Deletion | Chr8 | 6204619 | 6204622 | 3 | |
| Deletion | Chr11 | 6331001 | 6331004 | 3 | |
| Deletion | Chr2 | 7755452 | 7755453 | 1 | |
| Deletion | Chr8 | 9380282 | 9380298 | 16 | |
| Deletion | Chr5 | 11819404 | 11819474 | 70 | |
| Deletion | Chr12 | 13000109 | 13000110 | 1 | LOC_Os12g22990 |
| Deletion | Chr3 | 13895001 | 13932000 | 37000 | 6 |
| Deletion | Chr1 | 14130331 | 14130340 | 9 | |
| Deletion | Chr12 | 14245799 | 14245803 | 4 | LOC_Os12g24820 |
| Deletion | Chr3 | 14860760 | 14860766 | 6 | |
| Deletion | Chr9 | 15022001 | 15055000 | 33000 | 3 |
| Deletion | Chr2 | 16197870 | 16197891 | 21 | |
| Deletion | Chr8 | 16590537 | 16590541 | 4 | |
| Deletion | Chr8 | 16886269 | 16886270 | 1 | |
| Deletion | Chr8 | 17672482 | 17672485 | 3 | |
| Deletion | Chr6 | 20150250 | 20150251 | 1 | LOC_Os06g34670 |
| Deletion | Chr12 | 20377537 | 20377539 | 2 | |
| Deletion | Chr6 | 23679588 | 23679589 | 1 | |
| Deletion | Chr3 | 23863613 | 23863614 | 1 | |
| Deletion | Chr7 | 24092731 | 24092736 | 5 | |
| Deletion | Chr6 | 25269452 | 25269453 | 1 | LOC_Os06g42060 |
| Deletion | Chr11 | 26449452 | 26449453 | 1 | |
| Deletion | Chr11 | 27620495 | 27620512 | 17 | |
| Deletion | Chr1 | 35659740 | 35659742 | 2 | LOC_Os01g61630 |
| Deletion | Chr1 | 38799040 | 38799048 | 8 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr2 | 16949361 | 16949361 | 1 | LOC_Os02g28650 |
| Insertion | Chr2 | 25678710 | 25678751 | 42 | |
| Insertion | Chr3 | 31100647 | 31100648 | 2 | |
| Insertion | Chr5 | 29710421 | 29710423 | 3 | |
| Insertion | Chr6 | 2276270 | 2276272 | 3 | LOC_Os06g05130 |
| Insertion | Chr6 | 24610240 | 24610241 | 2 | |
| Insertion | Chr6 | 28887929 | 28887930 | 2 | |
| Insertion | Chr6 | 5543356 | 5543358 | 3 | |
| Insertion | Chr7 | 6862804 | 6862805 | 2 | |
| Insertion | Chr8 | 8941178 | 8941178 | 1 | |
| Insertion | Chr9 | 6558056 | 6558057 | 2 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 1853130 | 2106164 | 2 |
| Inversion | Chr1 | 1994304 | 2106158 | 2 |
| Inversion | Chr7 | 7169400 | 7200394 | 2 |
| Inversion | Chr7 | 7169465 | 7200396 | 2 |
| Inversion | Chr6 | 11043998 | 11545587 | |
| Inversion | Chr5 | 11445139 | 11629627 | |
| Inversion | Chr3 | 13895524 | 13955315 | |
| Inversion | Chr3 | 13932253 | 13955325 | LOC_Os03g24450 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 2967476 | Chr1 | 23436212 | LOC_Os04g05780 |
| Translocation | Chr11 | 12597110 | Chr9 | 1340905 | |
| Translocation | Chr10 | 20577014 | Chr7 | 22025409 | LOC_Os07g36760 |
| Translocation | Chr6 | 21657580 | Chr4 | 17923468 | LOC_Os06g36770 |
| Translocation | Chr7 | 22011588 | Chr5 | 20489094 | 2 |
