Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1665-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1665-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 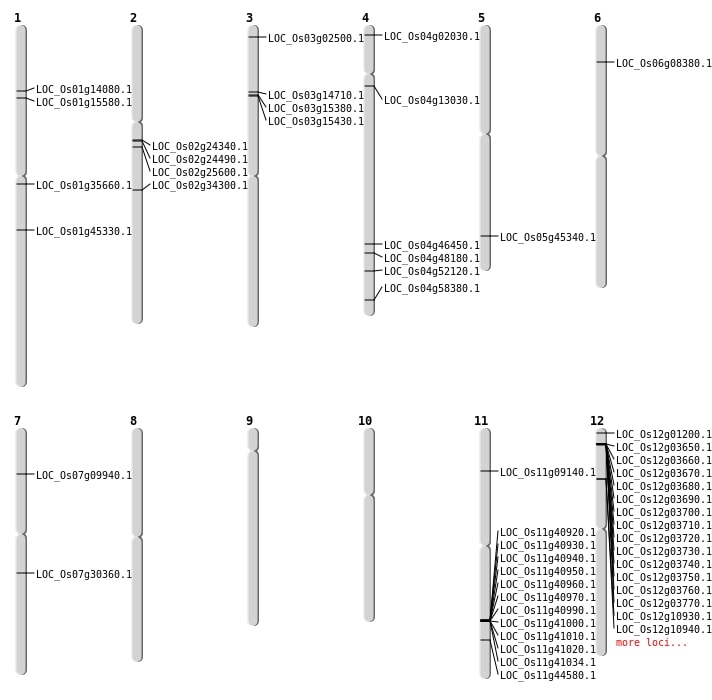
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 16089838 | T-A | HET | INTRON | |
| SBS | Chr1 | 16089840 | C-A | HET | INTRON | |
| SBS | Chr1 | 23686120 | C-T | HET | INTERGENIC | |
| SBS | Chr1 | 25203049 | T-A | HET | INTERGENIC | |
| SBS | Chr1 | 4088199 | T-C | HET | INTERGENIC | |
| SBS | Chr1 | 8681983 | G-A | HET | INTERGENIC | |
| SBS | Chr11 | 19916994 | T-C | HET | INTERGENIC | |
| SBS | Chr11 | 26954608 | A-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44580 |
| SBS | Chr12 | 21838571 | T-A | HET | INTERGENIC | |
| SBS | Chr12 | 2271277 | G-T | Homo | INTERGENIC | |
| SBS | Chr12 | 3739101 | G-C | Homo | INTERGENIC | |
| SBS | Chr2 | 20526845 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g34300 |
| SBS | Chr2 | 26438071 | C-G | Homo | INTERGENIC | |
| SBS | Chr3 | 25981579 | G-A | Homo | INTRON | |
| SBS | Chr4 | 1371867 | T-A | Homo | INTERGENIC | |
| SBS | Chr4 | 28689390 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g48180 |
| SBS | Chr4 | 627489 | G-A | Homo | INTRON | |
| SBS | Chr5 | 13406371 | A-G | HET | INTRON | |
| SBS | Chr5 | 29886601 | T-A | Homo | INTRON | |
| SBS | Chr5 | 3471914 | A-T | Homo | INTERGENIC | |
| SBS | Chr5 | 5527300 | G-T | Homo | INTERGENIC | |
| SBS | Chr6 | 16698171 | T-C | HET | INTRON | |
| SBS | Chr6 | 16934678 | G-T | HET | INTRON | |
| SBS | Chr6 | 24977175 | T-A | Homo | INTRON | |
| SBS | Chr6 | 7050124 | T-C | HET | SYNONYMOUS_CODING | |
| SBS | Chr7 | 10278506 | T-A | Homo | INTERGENIC | |
| SBS | Chr7 | 11317925 | A-G | HET | INTERGENIC | |
| SBS | Chr7 | 17965748 | G-T | Homo | STOP_GAINED | LOC_Os07g30360 |
| SBS | Chr7 | 21883411 | G-A | HET | INTERGENIC | |
| SBS | Chr7 | 22350225 | C-T | HET | INTERGENIC | |
| SBS | Chr7 | 5293781 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g09940 |
| SBS | Chr8 | 17462447 | C-T | Homo | INTERGENIC | |
| SBS | Chr8 | 19346982 | C-T | HET | INTERGENIC | |
| SBS | Chr8 | 8975213 | A-C | Homo | INTERGENIC | |
| SBS | Chr9 | 21407738 | C-T | HET | INTERGENIC | |
| SBS | Chr9 | 7197707 | A-T | HET | INTERGENIC |
Deletions: 36
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 116843 | 116858 | 15 | LOC_Os12g01200 |
| Deletion | Chr4 | 639892 | 639893 | 1 | LOC_Os04g02030 |
| Deletion | Chr7 | 1194231 | 1194236 | 5 | |
| Deletion | Chr12 | 1464001 | 1534000 | 70000 | 13 |
| Deletion | Chr9 | 1926812 | 1926813 | 1 | |
| Deletion | Chr10 | 2165624 | 2165626 | 2 | |
| Deletion | Chr11 | 3584307 | 3584308 | 1 | |
| Deletion | Chr8 | 3586675 | 3586676 | 1 | |
| Deletion | Chr4 | 4455893 | 4455912 | 19 | |
| Deletion | Chr1 | 5626328 | 5626331 | 3 | |
| Deletion | Chr12 | 5906001 | 6520000 | 614000 | 94 |
| Deletion | Chr9 | 6544883 | 6544895 | 12 | |
| Deletion | Chr4 | 7716593 | 7716604 | 11 | |
| Deletion | Chr1 | 7882546 | 7882553 | 7 | LOC_Os01g14080 |
| Deletion | Chr10 | 7991097 | 7991101 | 4 | |
| Deletion | Chr12 | 8314714 | 8314715 | 1 | |
| Deletion | Chr8 | 8889012 | 8889013 | 1 | |
| Deletion | Chr9 | 9305056 | 9305068 | 12 | |
| Deletion | Chr8 | 11247358 | 11247369 | 11 | |
| Deletion | Chr7 | 11859978 | 11859979 | 1 | |
| Deletion | Chr3 | 14088267 | 14088271 | 4 | |
| Deletion | Chr12 | 17167341 | 17167342 | 1 | |
| Deletion | Chr11 | 18808766 | 18808774 | 8 | |
| Deletion | Chr9 | 19297946 | 19297949 | 3 | |
| Deletion | Chr1 | 19751626 | 19751627 | 1 | LOC_Os01g35660 |
| Deletion | Chr1 | 22462127 | 22462128 | 1 | |
| Deletion | Chr2 | 24090639 | 24090645 | 6 | |
| Deletion | Chr11 | 24499001 | 24566000 | 67000 | 11 |
| Deletion | Chr12 | 24971710 | 24971711 | 1 | |
| Deletion | Chr5 | 24986899 | 24986902 | 3 | |
| Deletion | Chr5 | 26304361 | 26304365 | 4 | LOC_Os05g45340 |
| Deletion | Chr2 | 28127751 | 28127752 | 1 | |
| Deletion | Chr1 | 28376017 | 28376021 | 4 | |
| Deletion | Chr4 | 30952523 | 30952531 | 8 | LOC_Os04g52120 |
| Deletion | Chr4 | 32470343 | 32470358 | 15 | |
| Deletion | Chr4 | 33935344 | 33935352 | 8 |
Insertions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 17956443 | 17956443 | 1 | |
| Insertion | Chr1 | 21313909 | 21313909 | 1 | |
| Insertion | Chr1 | 2410544 | 2410544 | 1 | |
| Insertion | Chr1 | 2504910 | 2504951 | 42 | |
| Insertion | Chr1 | 25730141 | 25730153 | 13 | LOC_Os01g45330 |
| Insertion | Chr1 | 29199100 | 29199101 | 2 | |
| Insertion | Chr1 | 6640664 | 6640673 | 10 | |
| Insertion | Chr1 | 8751109 | 8751114 | 6 | LOC_Os01g15580 |
| Insertion | Chr10 | 6722414 | 6722437 | 24 | |
| Insertion | Chr11 | 17890746 | 17890787 | 42 | |
| Insertion | Chr11 | 4866717 | 4866718 | 2 | LOC_Os11g09140 |
| Insertion | Chr2 | 4256994 | 4256995 | 2 | |
| Insertion | Chr3 | 1061044 | 1061055 | 12 | |
| Insertion | Chr3 | 1240278 | 1240278 | 1 | |
| Insertion | Chr3 | 1257473 | 1257474 | 2 | |
| Insertion | Chr3 | 921435 | 921477 | 43 | LOC_Os03g02500 |
| Insertion | Chr4 | 26425841 | 26425841 | 1 | |
| Insertion | Chr4 | 7188616 | 7188679 | 64 | LOC_Os04g13030 |
| Insertion | Chr4 | 8338683 | 8338683 | 1 | |
| Insertion | Chr6 | 30781507 | 30781507 | 1 | |
| Insertion | Chr7 | 14959981 | 14959982 | 2 | |
| Insertion | Chr7 | 29629256 | 29629257 | 2 |
Inversions: 9
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 4043063 | 4068268 | LOC_Os06g08380 |
| Inversion | Chr6 | 4046889 | 4068270 | LOC_Os06g08380 |
| Inversion | Chr3 | 7999182 | 8414338 | 2 |
| Inversion | Chr3 | 8415122 | 8439528 | LOC_Os03g15430 |
| Inversion | Chr12 | 11968690 | 11987546 | |
| Inversion | Chr2 | 14121142 | 14964763 | 2 |
| Inversion | Chr2 | 14189569 | 14964757 | 2 |
| Inversion | Chr4 | 17939830 | 27557393 | 2 |
| Inversion | Chr4 | 34733603 | 34785758 | LOC_Os04g58380 |
No Translocation
Tandem duplications: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Tandem Duplication | Chr3 | 7999191 | 7999191 | 0 | LOC_Os03g14710 |
