Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1668-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1668-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 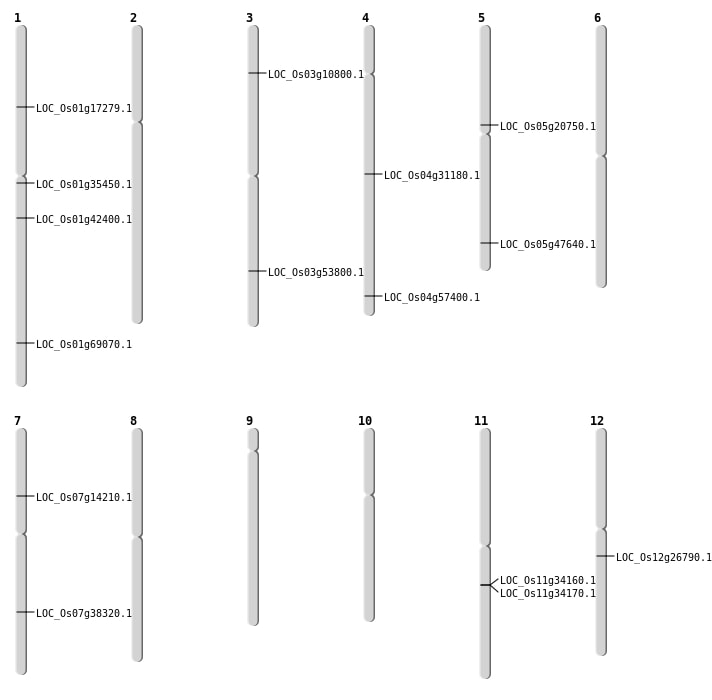
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 1075552 | G-A | HET | ||
| SBS | Chr1 | 19616858 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g35450 |
| SBS | Chr1 | 22498714 | C-T | Homo | ||
| SBS | Chr1 | 22663640 | C-T | HET | ||
| SBS | Chr1 | 33991436 | G-A | HET | ||
| SBS | Chr1 | 34083549 | A-C | HET | ||
| SBS | Chr10 | 5924411 | T-G | HET | ||
| SBS | Chr11 | 17531808 | G-A | Homo | ||
| SBS | Chr11 | 4370091 | A-G | Homo | ||
| SBS | Chr2 | 13868016 | G-T | HET | ||
| SBS | Chr2 | 16818781 | G-A | HET | ||
| SBS | Chr2 | 31055631 | T-A | Homo | ||
| SBS | Chr2 | 335824 | T-A | HET | ||
| SBS | Chr3 | 13938504 | A-G | Homo | ||
| SBS | Chr3 | 25939135 | C-G | HET | ||
| SBS | Chr3 | 29588685 | G-A | HET | ||
| SBS | Chr3 | 5532446 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g10800 |
| SBS | Chr4 | 34157473 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g57400 |
| SBS | Chr5 | 12178208 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g20750 |
| SBS | Chr5 | 15700821 | T-C | HET | ||
| SBS | Chr5 | 7536407 | A-T | HET | ||
| SBS | Chr6 | 3716104 | T-A | Homo | ||
| SBS | Chr7 | 23020871 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g38320 |
| SBS | Chr7 | 8099578 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os07g14210 |
| SBS | Chr8 | 2001589 | C-A | Homo | ||
| SBS | Chr9 | 16648204 | C-T | HET | ||
| SBS | Chr9 | 19867417 | C-T | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 2163415 | 2163419 | 4 | |
| Deletion | Chr11 | 4373603 | 4373606 | 3 | |
| Deletion | Chr2 | 4513234 | 4513236 | 2 | |
| Deletion | Chr9 | 5084012 | 5084013 | 1 | |
| Deletion | Chr9 | 5472348 | 5472362 | 14 | |
| Deletion | Chr1 | 9946691 | 9946699 | 8 | LOC_Os01g17279 |
| Deletion | Chr11 | 10002733 | 10002734 | 1 | |
| Deletion | Chr4 | 10422025 | 10422026 | 1 | |
| Deletion | Chr12 | 15699181 | 15699188 | 7 | LOC_Os12g26790 |
| Deletion | Chr1 | 17679312 | 17679313 | 1 | |
| Deletion | Chr10 | 17922774 | 17922778 | 4 | |
| Deletion | Chr1 | 19509866 | 19509870 | 4 | |
| Deletion | Chr9 | 19832451 | 19832455 | 4 | |
| Deletion | Chr2 | 20486674 | 20486680 | 6 | |
| Deletion | Chr11 | 21727809 | 21727826 | 17 | |
| Deletion | Chr1 | 23974270 | 23974285 | 15 | |
| Deletion | Chr2 | 23980292 | 23980299 | 7 | |
| Deletion | Chr1 | 24105065 | 24105078 | 13 | LOC_Os01g42400 |
| Deletion | Chr6 | 29078510 | 29078527 | 17 | |
| Deletion | Chr6 | 29458425 | 29458426 | 1 | |
| Deletion | Chr6 | 30346165 | 30346175 | 10 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 12114282 | 12114334 | 53 | |
| Insertion | Chr9 | 21644056 | 21644059 | 4 |
Inversions: 13
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 308352 | 308710 | |
| Inversion | Chr1 | 1627278 | 1627510 | |
| Inversion | Chr9 | 6207433 | 6207869 | |
| Inversion | Chr3 | 12800358 | 12800552 | |
| Inversion | Chr11 | 20005327 | 20005596 | 2 |
| Inversion | Chr2 | 25636605 | 25636812 | |
| Inversion | Chr4 | 26440931 | 26441396 | |
| Inversion | Chr5 | 27120871 | 27290825 | 2 |
| Inversion | Chr5 | 27120927 | 27290834 | 2 |
| Inversion | Chr3 | 30629570 | 30855290 | 2 |
| Inversion | Chr3 | 30629617 | 30855291 | 2 |
| Inversion | Chr2 | 35245427 | 35245616 | |
| Inversion | Chr1 | 40141433 | 40141592 | LOC_Os01g69070 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 5049974 | Chr4 | 18657034 | 2 |
| Translocation | Chr3 | 9138528 | Chr2 | 31054320 | |
| Translocation | Chr3 | 9138534 | Chr2 | 31054377 | |
| Translocation | Chr9 | 11307386 | Chr6 | 22141205 |
