Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1675-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1675-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 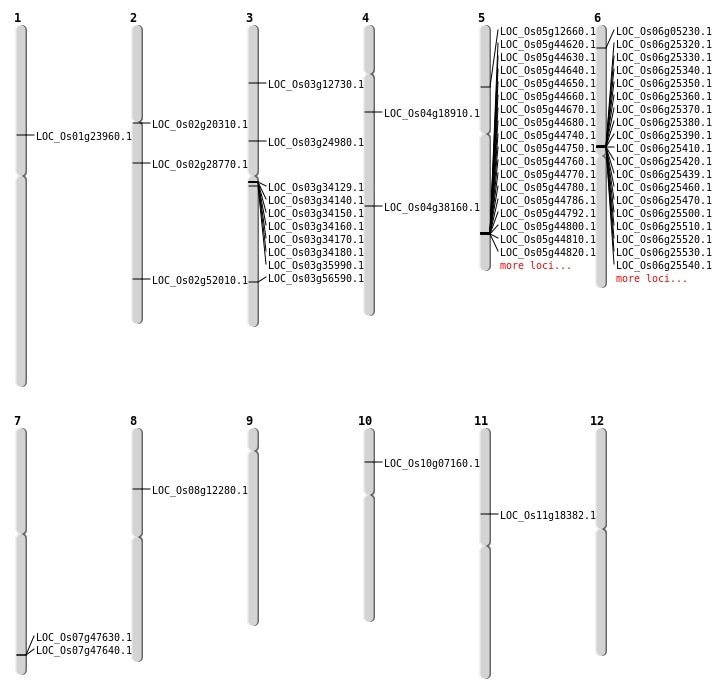
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11123062 | T-C | Homo | ||
| SBS | Chr1 | 13410720 | G-A | HET | ||
| SBS | Chr1 | 2549452 | C-T | Homo | ||
| SBS | Chr10 | 1095189 | T-C | HET | ||
| SBS | Chr10 | 18307964 | G-A | HET | ||
| SBS | Chr10 | 8441129 | C-G | HET | ||
| SBS | Chr11 | 10379172 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g18382 |
| SBS | Chr11 | 18617774 | G-T | Homo | ||
| SBS | Chr11 | 8252707 | C-G | HET | ||
| SBS | Chr2 | 15857991 | G-T | Homo | ||
| SBS | Chr2 | 20017975 | G-A | Homo | ||
| SBS | Chr2 | 28892913 | C-T | Homo | ||
| SBS | Chr2 | 31840754 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g52010 |
| SBS | Chr3 | 6805782 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g12730 |
| SBS | Chr4 | 22710374 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g38160 |
| SBS | Chr4 | 31317162 | T-C | HET | ||
| SBS | Chr4 | 4205110 | G-T | Homo | ||
| SBS | Chr4 | 4205111 | C-T | Homo | ||
| SBS | Chr5 | 2592313 | A-G | Homo | ||
| SBS | Chr6 | 19552656 | C-T | HET | ||
| SBS | Chr6 | 19852834 | G-A | HET | ||
| SBS | Chr6 | 2349528 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g05230 |
| SBS | Chr6 | 9413862 | G-A | HET | ||
| SBS | Chr6 | 9413863 | C-T | HET | ||
| SBS | Chr7 | 15078794 | T-A | HET | ||
| SBS | Chr8 | 23425091 | G-A | HET | ||
| SBS | Chr9 | 5365438 | T-G | HET |
Deletions: 13
Insertions: 7
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 11114415 | 11114415 | 1 | |
| Insertion | Chr11 | 13493795 | 13493795 | 1 | |
| Insertion | Chr4 | 10516215 | 10516215 | 1 | LOC_Os04g18910 |
| Insertion | Chr4 | 3903077 | 3903077 | 1 | |
| Insertion | Chr7 | 9903210 | 9903210 | 1 | |
| Insertion | Chr8 | 5022341 | 5022343 | 3 | |
| Insertion | Chr8 | 6945205 | 6945205 | 1 |
Inversions: 8
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 7229324 | 7457437 | LOC_Os08g12280 |
| Inversion | Chr1 | 13481437 | 13481850 | LOC_Os01g23960 |
| Inversion | Chr9 | 16371508 | 16371866 | |
| Inversion | Chr5 | 17747157 | 18146585 | |
| Inversion | Chr6 | 19623967 | 19997981 | |
| Inversion | Chr6 | 19624463 | 19842432 | 2 |
| Inversion | Chr3 | 32077467 | 32238260 | 2 |
| Inversion | Chr3 | 32077485 | 32238265 | 2 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1930158 | Chr4 | 25011835 | |
| Translocation | Chr11 | 1930500 | Chr4 | 25011834 | |
| Translocation | Chr10 | 3761462 | Chr2 | 17525907 | LOC_Os10g07160 |
| Translocation | Chr5 | 7269477 | Chr3 | 19922722 | LOC_Os05g12660 |
| Translocation | Chr7 | 7319153 | Chr5 | 26011591 | 2 |
| Translocation | Chr9 | 9016527 | Chr7 | 28474575 | 2 |
| Translocation | Chr9 | 9016539 | Chr7 | 28473838 | 2 |
| Translocation | Chr3 | 14255790 | Chr2 | 17025208 | 2 |
| Translocation | Chr6 | 17871915 | Chr2 | 11966173 | 2 |
| Translocation | Chr4 | 17926761 | Chr3 | 9616514 | |
| Translocation | Chr3 | 19964935 | Chr1 | 40083673 | LOC_Os03g35990 |
| Translocation | Chr10 | 20572652 | Chr1 | 41377868 | |
| Translocation | Chr7 | 23329929 | Chr5 | 26011591 | 2 |
| Translocation | Chr7 | 23329955 | Chr5 | 26011789 | 2 |
