Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1690-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1690-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 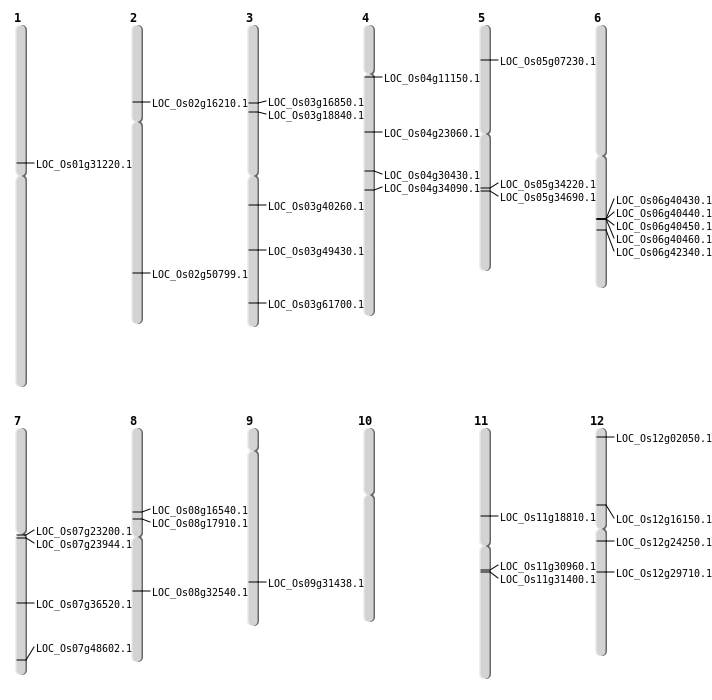
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 23705725 | A-G | HET | ||
| SBS | Chr11 | 14780607 | T-C | HET | ||
| SBS | Chr11 | 23083481 | A-T | HET | ||
| SBS | Chr12 | 13198287 | T-A | HET | ||
| SBS | Chr12 | 2192875 | G-T | HET | ||
| SBS | Chr12 | 9221276 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g16150 |
| SBS | Chr2 | 10782180 | T-C | HET | ||
| SBS | Chr2 | 17754896 | G-A | HET | ||
| SBS | Chr2 | 32278526 | T-C | HET | ||
| SBS | Chr2 | 5510396 | A-T | HET | ||
| SBS | Chr2 | 7237161 | A-G | HET | ||
| SBS | Chr2 | 9213368 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16210 |
| SBS | Chr3 | 10280830 | G-T | HET | ||
| SBS | Chr3 | 23914236 | G-A | HET | ||
| SBS | Chr3 | 9360203 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os03g16850 |
| SBS | Chr4 | 19584222 | A-C | HET | ||
| SBS | Chr4 | 24928817 | T-C | HET | ||
| SBS | Chr4 | 4502866 | T-C | HET | ||
| SBS | Chr5 | 23780553 | G-A | HET | ||
| SBS | Chr5 | 29715159 | T-G | HET | ||
| SBS | Chr6 | 13937349 | T-G | HET | ||
| SBS | Chr6 | 9536934 | C-T | HET | ||
| SBS | Chr7 | 13198919 | G-A | HET | ||
| SBS | Chr7 | 15523331 | C-T | Homo | ||
| SBS | Chr7 | 25880579 | G-A | HET | ||
| SBS | Chr9 | 17333540 | A-T | HET |
Deletions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 3907669 | 3907671 | 2 | |
| Deletion | Chr3 | 6024347 | 6024349 | 2 | |
| Deletion | Chr7 | 8794163 | 8794164 | 1 | |
| Deletion | Chr12 | 17733001 | 17773000 | 39999 | LOC_Os12g29710 |
| Deletion | Chr6 | 24078001 | 24105000 | 26999 | 4 |
| Deletion | Chr3 | 28141320 | 28141321 | 1 | LOC_Os03g49430 |
Insertions: 2
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 18716593 | 18716593 | 1 | |
| Insertion | Chr6 | 5048099 | 5048100 | 2 |
Inversions: 26
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 610669 | 610886 | LOC_Os12g02050 |
| Inversion | Chr12 | 2017049 | 2017359 | |
| Inversion | Chr1 | 2392978 | 3356117 | |
| Inversion | Chr5 | 3825592 | 3825723 | LOC_Os05g07230 |
| Inversion | Chr5 | 4911557 | 4911702 | |
| Inversion | Chr5 | 5347977 | 5348271 | |
| Inversion | Chr3 | 10556668 | 10556934 | LOC_Os03g18840 |
| Inversion | Chr8 | 10778094 | 10778217 | |
| Inversion | Chr7 | 13077616 | 13535868 | 2 |
| Inversion | Chr7 | 13077628 | 13540219 | 2 |
| Inversion | Chr12 | 13827822 | 13828124 | LOC_Os12g24250 |
| Inversion | Chr10 | 14428791 | 14429004 | LOC_Os10g27340 |
| Inversion | Chr1 | 16534585 | 16534692 | |
| Inversion | Chr1 | 17067499 | 17067945 | LOC_Os01g31220 |
| Inversion | Chr4 | 18172470 | 18172589 | LOC_Os04g30430 |
| Inversion | Chr9 | 18920938 | 18921266 | LOC_Os09g31438 |
| Inversion | Chr5 | 20259586 | 20259886 | LOC_Os05g34220 |
| Inversion | Chr5 | 20583683 | 20584064 | 2 |
| Inversion | Chr7 | 21726548 | 21837021 | 2 |
| Inversion | Chr7 | 21726593 | 21837029 | 2 |
| Inversion | Chr11 | 22299377 | 22299939 | |
| Inversion | Chr3 | 22379197 | 22379352 | LOC_Os03g40260 |
| Inversion | Chr2 | 22605796 | 22606007 | |
| Inversion | Chr7 | 28923896 | 28924269 | |
| Inversion | Chr7 | 29098794 | 29099004 | LOC_Os07g48602 |
| Inversion | Chr4 | 33189140 | 33189575 |
Translocations: 14
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 1518989 | Chr6 | 25438347 | 2 |
| Translocation | Chr4 | 6061531 | Chr3 | 34971581 | 2 |
| Translocation | Chr4 | 6062785 | Chr3 | 34971586 | 2 |
| Translocation | Chr12 | 8500490 | Chr6 | 11334665 | |
| Translocation | Chr8 | 10124260 | Chr6 | 15690600 | LOC_Os08g16540 |
| Translocation | Chr9 | 10353230 | Chr5 | 14533759 | |
| Translocation | Chr9 | 10353250 | Chr8 | 18506834 | |
| Translocation | Chr11 | 10672582 | Chr8 | 10989191 | 2 |
| Translocation | Chr9 | 15029389 | Chr2 | 31025674 | 2 |
| Translocation | Chr10 | 16453731 | Chr6 | 24106238 | 2 |
| Translocation | Chr11 | 18012810 | Chr4 | 13099600 | 2 |
| Translocation | Chr11 | 18321774 | Chr10 | 6935147 | LOC_Os11g31400 |
| Translocation | Chr11 | 18322578 | Chr4 | 20652378 | 2 |
| Translocation | Chr8 | 20159704 | Chr5 | 14039973 | LOC_Os08g32540 |
