Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1697-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1697-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 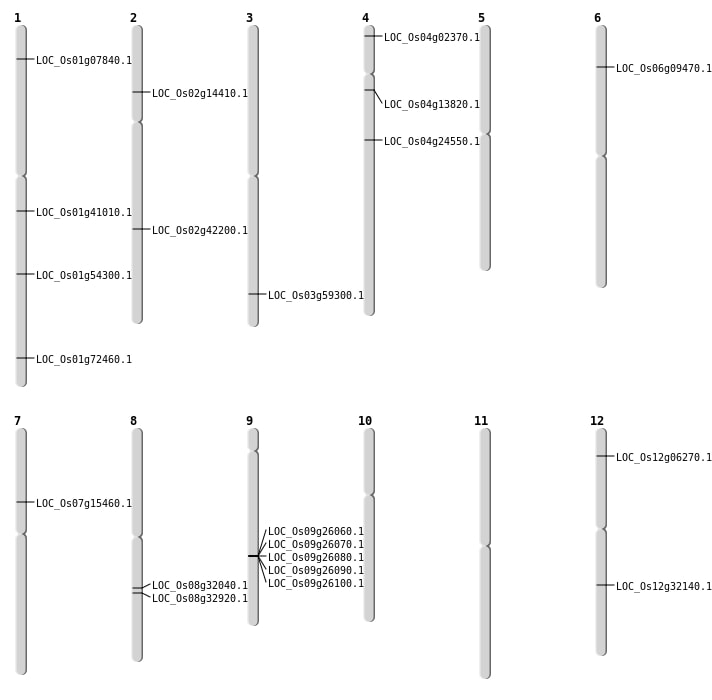
|
| Variant Information |
|---|
Single base substitutions: 48
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11401579 | A-T | HET | ||
| SBS | Chr1 | 15288532 | T-A | HET | ||
| SBS | Chr1 | 16117194 | A-T | HET | ||
| SBS | Chr1 | 21561525 | A-T | HET | ||
| SBS | Chr1 | 3766090 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g07840 |
| SBS | Chr1 | 40685653 | T-A | Homo | ||
| SBS | Chr10 | 5486005 | T-A | Homo | ||
| SBS | Chr11 | 15732030 | G-C | HET | ||
| SBS | Chr11 | 4316109 | G-A | HET | ||
| SBS | Chr12 | 15338502 | T-G | HET | ||
| SBS | Chr12 | 19383942 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g32140 |
| SBS | Chr12 | 19383943 | G-T | HET | ||
| SBS | Chr12 | 19383944 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g32140 |
| SBS | Chr12 | 19520105 | A-T | HET | ||
| SBS | Chr12 | 25473382 | T-A | HET | ||
| SBS | Chr12 | 8954890 | G-T | Homo | ||
| SBS | Chr2 | 16201766 | A-T | HET | ||
| SBS | Chr2 | 20660831 | T-C | HET | ||
| SBS | Chr2 | 22289326 | A-T | HET | ||
| SBS | Chr2 | 31190242 | A-C | HET | ||
| SBS | Chr2 | 7911521 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14410 |
| SBS | Chr3 | 25374096 | T-G | HET | ||
| SBS | Chr3 | 30192078 | G-A | HET | ||
| SBS | Chr4 | 10792584 | C-T | Homo | ||
| SBS | Chr4 | 7710204 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13820 |
| SBS | Chr5 | 15419560 | C-G | HET | ||
| SBS | Chr6 | 11367176 | G-C | HET | ||
| SBS | Chr6 | 143738 | G-T | HET | ||
| SBS | Chr6 | 1506600 | G-A | HET | ||
| SBS | Chr6 | 1506601 | A-T | HET | ||
| SBS | Chr6 | 23354319 | A-T | HET | ||
| SBS | Chr6 | 3112379 | C-T | HET | ||
| SBS | Chr7 | 10211462 | C-T | HET | ||
| SBS | Chr7 | 14026266 | C-T | Homo | ||
| SBS | Chr7 | 8970228 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g15460 |
| SBS | Chr7 | 9334975 | T-C | HET | ||
| SBS | Chr8 | 11012988 | C-T | HET | ||
| SBS | Chr8 | 12833719 | G-A | HET | ||
| SBS | Chr8 | 15815596 | C-T | HET | ||
| SBS | Chr8 | 19869715 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g32040 |
| SBS | Chr8 | 23680304 | A-G | HET | ||
| SBS | Chr8 | 27377576 | G-A | HET | ||
| SBS | Chr8 | 28197871 | A-C | HET | ||
| SBS | Chr9 | 12879232 | A-T | Homo | ||
| SBS | Chr9 | 15022546 | G-A | HET | ||
| SBS | Chr9 | 2149952 | C-T | HET | ||
| SBS | Chr9 | 2151298 | A-C | HET | ||
| SBS | Chr9 | 4743356 | T-G | HET |
Deletions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 3409287 | 3409289 | 2 | |
| Deletion | Chr9 | 9279696 | 9279703 | 7 | |
| Deletion | Chr4 | 14081001 | 14091000 | 9999 | LOC_Os04g24550 |
| Deletion | Chr9 | 15679001 | 15707000 | 27999 | 5 |
| Deletion | Chr12 | 19081555 | 19081556 | 1 | |
| Deletion | Chr6 | 24100540 | 24100542 | 2 |
Insertions: 8
Inversions: 13
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr6 | 1215569 | 1215765 | |
| Inversion | Chr7 | 9398785 | 9399036 | |
| Inversion | Chr11 | 15547266 | 16038680 | |
| Inversion | Chr11 | 15547272 | 16038688 | |
| Inversion | Chr9 | 16806546 | 16806879 | |
| Inversion | Chr9 | 16881027 | 16881378 | |
| Inversion | Chr8 | 20416961 | 20417322 | LOC_Os08g32920 |
| Inversion | Chr1 | 23225388 | 23225651 | LOC_Os01g41010 |
| Inversion | Chr7 | 23677160 | 23677479 | |
| Inversion | Chr4 | 29093816 | 29094018 | |
| Inversion | Chr1 | 31253934 | 31254063 | LOC_Os01g54300 |
| Inversion | Chr3 | 33766463 | 33766671 | LOC_Os03g59300 |
| Inversion | Chr1 | 42017897 | 42018151 | 2 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 2984774 | Chr9 | 2136220 | LOC_Os12g06270 |
| Translocation | Chr12 | 2985475 | Chr9 | 2136209 | LOC_Os12g06270 |
| Translocation | Chr6 | 4820846 | Chr4 | 848440 | 2 |
| Translocation | Chr11 | 7098110 | Chr6 | 1748200 | |
| Translocation | Chr6 | 9165160 | Chr3 | 14831130 | |
| Translocation | Chr5 | 9977861 | Chr2 | 14658117 | |
| Translocation | Chr7 | 15462310 | Chr6 | 24777049 | |
| Translocation | Chr7 | 19555902 | Chr2 | 25382838 | 2 |
