Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1707-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1707-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 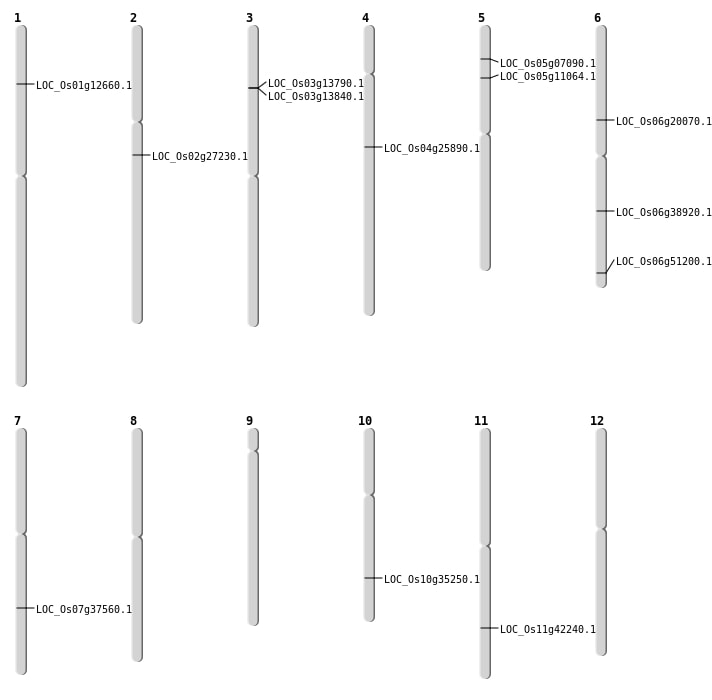
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 24292437 | C-T | Homo | ||
| SBS | Chr1 | 3237224 | A-G | HET | ||
| SBS | Chr1 | 38209651 | C-T | HET | ||
| SBS | Chr1 | 7661998 | G-A | Homo | ||
| SBS | Chr10 | 12525436 | C-T | HET | ||
| SBS | Chr10 | 13865606 | C-T | HET | ||
| SBS | Chr10 | 17518125 | A-G | HET | ||
| SBS | Chr10 | 18838180 | G-A | HET | ||
| SBS | Chr11 | 1180964 | A-C | HET | ||
| SBS | Chr11 | 14036685 | A-C | HET | ||
| SBS | Chr11 | 14100823 | T-C | Homo | ||
| SBS | Chr11 | 20783228 | T-A | Homo | ||
| SBS | Chr11 | 25441397 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g42240 |
| SBS | Chr11 | 9600557 | G-T | HET | ||
| SBS | Chr12 | 11500530 | A-G | Homo | ||
| SBS | Chr12 | 16992700 | G-A | HET | ||
| SBS | Chr12 | 17384071 | A-C | HET | ||
| SBS | Chr12 | 19792060 | G-A | HET | ||
| SBS | Chr12 | 19792104 | T-A | HET | ||
| SBS | Chr12 | 26067283 | C-A | HET | ||
| SBS | Chr2 | 11990543 | G-A | HET | ||
| SBS | Chr2 | 16022315 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g27230 |
| SBS | Chr2 | 32320025 | G-C | Homo | ||
| SBS | Chr2 | 34901839 | T-C | Homo | ||
| SBS | Chr3 | 13281132 | G-T | HET | ||
| SBS | Chr3 | 29220457 | A-G | HET | ||
| SBS | Chr3 | 30759476 | T-G | HET | ||
| SBS | Chr3 | 36180413 | G-A | HET | ||
| SBS | Chr3 | 7129959 | C-T | Homo | ||
| SBS | Chr4 | 10541021 | T-C | HET | ||
| SBS | Chr4 | 13888214 | G-A | HET | ||
| SBS | Chr4 | 15057041 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g25890 |
| SBS | Chr4 | 20930398 | G-A | Homo | ||
| SBS | Chr4 | 27441010 | G-T | Homo | ||
| SBS | Chr4 | 31335478 | G-A | Homo | ||
| SBS | Chr4 | 31335479 | A-T | Homo | ||
| SBS | Chr5 | 13873790 | A-C | HET | ||
| SBS | Chr5 | 1965199 | A-G | Homo | ||
| SBS | Chr5 | 20860796 | C-T | Homo | ||
| SBS | Chr5 | 23474715 | C-T | HET | ||
| SBS | Chr5 | 25725566 | T-C | Homo | ||
| SBS | Chr5 | 3733716 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os05g07090 |
| SBS | Chr6 | 11510531 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g20070 |
| SBS | Chr6 | 21858413 | T-A | HET | ||
| SBS | Chr6 | 23731232 | G-T | HET | ||
| SBS | Chr6 | 25369291 | C-A | HET | ||
| SBS | Chr7 | 11936205 | C-T | Homo | ||
| SBS | Chr7 | 22489357 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g37560 |
| SBS | Chr7 | 22489359 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g37560 |
| SBS | Chr7 | 23482033 | T-A | HET | ||
| SBS | Chr7 | 6568810 | C-T | Homo | ||
| SBS | Chr7 | 7402342 | C-T | Homo | ||
| SBS | Chr8 | 20488083 | A-G | HET | ||
| SBS | Chr8 | 20521717 | G-A | HET | ||
| SBS | Chr8 | 26776968 | A-G | HET | ||
| SBS | Chr8 | 27837595 | C-T | HET | ||
| SBS | Chr9 | 9838272 | T-A | HET |
Deletions: 9
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr2 | 1489766 | 1489768 | 2 | |
| Deletion | Chr7 | 17632100 | 17632179 | 79 | |
| Deletion | Chr2 | 17801431 | 17801432 | 1 | |
| Deletion | Chr10 | 18838163 | 18838167 | 4 | LOC_Os10g35250 |
| Deletion | Chr12 | 19514034 | 19514035 | 1 | |
| Deletion | Chr3 | 20665289 | 20665290 | 1 | |
| Deletion | Chr3 | 27319486 | 27319489 | 3 | |
| Deletion | Chr4 | 31777307 | 31777308 | 1 | |
| Deletion | Chr1 | 32403175 | 32403179 | 4 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 26893886 | 26893889 | 4 | |
| Insertion | Chr5 | 17268170 | 17268171 | 2 | |
| Insertion | Chr5 | 21816867 | 21816867 | 1 | |
| Insertion | Chr6 | 23090979 | 23090979 | 1 | LOC_Os06g38920 |
Inversions: 6
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 729187 | Chr3 | 31273133 | |
| Translocation | Chr10 | 4089602 | Chr6 | 14634633 | |
| Translocation | Chr5 | 6200335 | Chr4 | 16995391 | LOC_Os05g11064 |
| Translocation | Chr7 | 9026353 | Chr2 | 7996989 | |
| Translocation | Chr11 | 17712239 | Chr6 | 30977071 | 2 |
| Translocation | Chr8 | 25425642 | Chr2 | 16056974 |
