Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1708-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1708-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 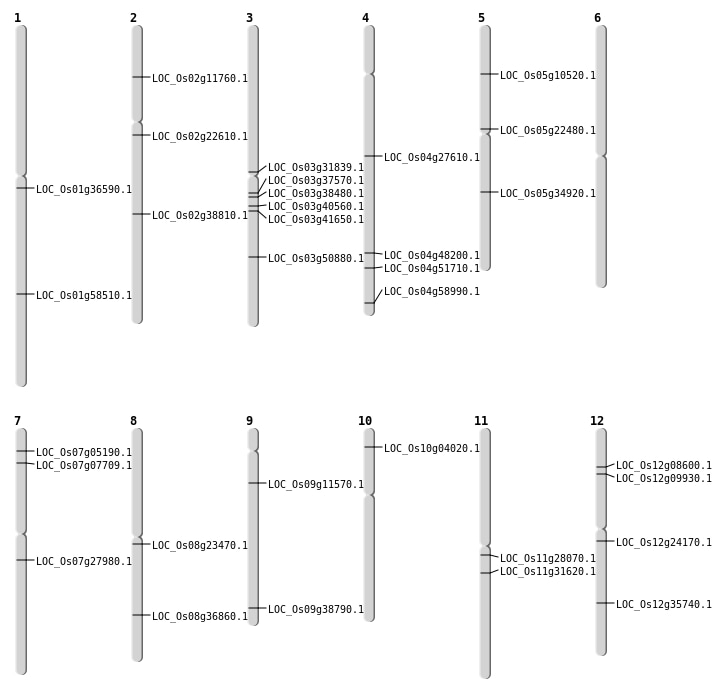
|
| Variant Information |
|---|
Single base substitutions: 23
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 7698780 | A-G | HET | ||
| SBS | Chr10 | 12480324 | G-A | Homo | ||
| SBS | Chr10 | 18046167 | G-A | Homo | ||
| SBS | Chr11 | 13265586 | G-T | HET | ||
| SBS | Chr11 | 25612310 | C-T | HET | ||
| SBS | Chr12 | 21762713 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g35740 |
| SBS | Chr12 | 23305993 | C-T | HET | ||
| SBS | Chr12 | 23305998 | T-C | HET | ||
| SBS | Chr2 | 29558680 | A-G | Homo | ||
| SBS | Chr2 | 9144896 | G-T | HET | ||
| SBS | Chr3 | 33962905 | C-T | HET | ||
| SBS | Chr4 | 26810722 | C-A | Homo | ||
| SBS | Chr4 | 27776567 | A-T | Homo | ||
| SBS | Chr4 | 35089772 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g58990 |
| SBS | Chr5 | 12763083 | G-A | HET | STOP_GAINED | LOC_Os05g22480 |
| SBS | Chr5 | 13966291 | G-A | HET | ||
| SBS | Chr5 | 6031046 | G-C | HET | ||
| SBS | Chr6 | 17721311 | G-C | HET | ||
| SBS | Chr6 | 20446734 | C-A | HET | ||
| SBS | Chr6 | 25534039 | T-G | HET | ||
| SBS | Chr7 | 7444214 | C-T | Homo | ||
| SBS | Chr9 | 10178731 | C-T | HET | ||
| SBS | Chr9 | 21959251 | C-G | HET |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 64116 | 64470 | 354 | |
| Deletion | Chr10 | 1839448 | 1839453 | 5 | LOC_Os10g04020 |
| Deletion | Chr4 | 9014565 | 9014574 | 9 | |
| Deletion | Chr1 | 10178376 | 10178503 | 127 | |
| Deletion | Chr4 | 11004137 | 11004138 | 1 | |
| Deletion | Chr9 | 13750392 | 13750393 | 1 | |
| Deletion | Chr8 | 18147092 | 18147093 | 1 | |
| Deletion | Chr3 | 18215232 | 18215239 | 7 | LOC_Os03g31839 |
| Deletion | Chr3 | 18476015 | 18476023 | 8 | |
| Deletion | Chr3 | 19793202 | 19793203 | 1 | |
| Deletion | Chr2 | 21912720 | 21912721 | 1 | |
| Deletion | Chr3 | 22555072 | 22555076 | 4 | LOC_Os03g40560 |
| Deletion | Chr11 | 27122724 | 27122731 | 7 |
Insertions: 5
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 1051332 | 1051332 | 1 | |
| Insertion | Chr11 | 12116424 | 12116465 | 42 | |
| Insertion | Chr11 | 22527025 | 22527027 | 3 | |
| Insertion | Chr2 | 1505963 | 1505963 | 1 | |
| Insertion | Chr4 | 16328224 | 16328224 | 1 | LOC_Os04g27610 |
Inversions: 41
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 2120046 | 2120161 | |
| Inversion | Chr7 | 2313561 | 2313694 | LOC_Os07g05190 |
| Inversion | Chr10 | 3671061 | 3671348 | |
| Inversion | Chr7 | 3872036 | 3872202 | 2 |
| Inversion | Chr6 | 3977391 | 3977654 | |
| Inversion | Chr12 | 4373470 | 4373593 | 2 |
| Inversion | Chr12 | 5259433 | 5259747 | LOC_Os12g09930 |
| Inversion | Chr5 | 5719501 | 5719629 | LOC_Os05g10520 |
| Inversion | Chr2 | 6070993 | 6071528 | LOC_Os02g11760 |
| Inversion | Chr9 | 6456635 | 6456835 | 2 |
| Inversion | Chr7 | 10127940 | 10128060 | |
| Inversion | Chr11 | 10818983 | 10819130 | |
| Inversion | Chr8 | 14212971 | 14213117 | LOC_Os08g23470 |
| Inversion | Chr9 | 14629105 | 14629430 | |
| Inversion | Chr4 | 14734654 | 14735008 | |
| Inversion | Chr11 | 16136813 | 16136935 | LOC_Os11g28070 |
| Inversion | Chr11 | 18489582 | 18489980 | LOC_Os11g31620 |
| Inversion | Chr3 | 20662744 | 20838936 | 2 |
| Inversion | Chr3 | 20662754 | 20838949 | 2 |
| Inversion | Chr5 | 20749799 | 20750068 | LOC_Os05g34920 |
| Inversion | Chr3 | 21358551 | 21358800 | LOC_Os03g38480 |
| Inversion | Chr9 | 22288866 | 22289061 | LOC_Os09g38790 |
| Inversion | Chr8 | 22375223 | 22375394 | |
| Inversion | Chr10 | 22852664 | 22852888 | |
| Inversion | Chr3 | 23153596 | 23153816 | LOC_Os03g41650 |
| Inversion | Chr8 | 23294104 | 23294278 | LOC_Os08g36860 |
| Inversion | Chr2 | 23464499 | 23464757 | LOC_Os02g38810 |
| Inversion | Chr7 | 25807905 | 25808358 | |
| Inversion | Chr2 | 25871475 | 25871594 | |
| Inversion | Chr12 | 26196173 | 26196399 | |
| Inversion | Chr4 | 27769290 | 27769703 | |
| Inversion | Chr4 | 28704340 | 28704521 | LOC_Os04g48200 |
| Inversion | Chr3 | 29069216 | 29082329 | LOC_Os03g50880 |
| Inversion | Chr3 | 29069218 | 29082343 | LOC_Os03g50880 |
| Inversion | Chr4 | 30644974 | 30645164 | LOC_Os04g51710 |
| Inversion | Chr3 | 33739916 | 33740214 | |
| Inversion | Chr1 | 33816225 | 33816606 | LOC_Os01g58510 |
| Inversion | Chr1 | 34099082 | 34099405 | |
| Inversion | Chr1 | 34166733 | 34166934 | |
| Inversion | Chr1 | 37424816 | 37425177 | |
| Inversion | Chr1 | 42054649 | 42055019 |
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 6098939 | Chr4 | 33550591 | |
| Translocation | Chr12 | 7608871 | Chr2 | 13477787 | 2 |
| Translocation | Chr12 | 11233290 | Chr1 | 11494819 | |
| Translocation | Chr12 | 13773249 | Chr4 | 714774 | LOC_Os12g24170 |
| Translocation | Chr7 | 16324864 | Chr3 | 22066452 | LOC_Os07g27980 |
| Translocation | Chr12 | 23130077 | Chr3 | 14084284 | |
| Translocation | Chr12 | 24065903 | Chr7 | 25224202 | |
| Translocation | Chr3 | 26025308 | Chr1 | 20308584 | 2 |
| Translocation | Chr12 | 27051819 | Chr7 | 5814191 |
