Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1739-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1739-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 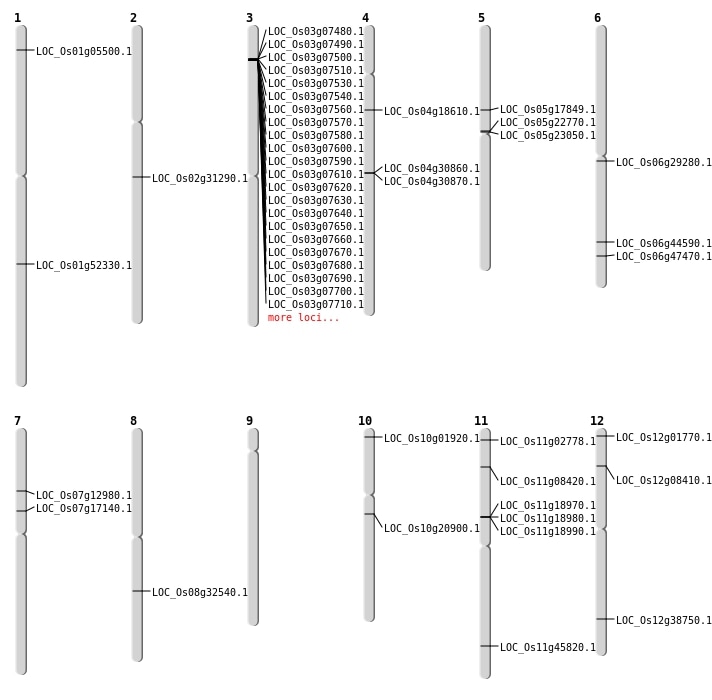
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11088201 | C-A | Homo | ||
| SBS | Chr1 | 35150823 | G-A | HET | ||
| SBS | Chr10 | 534980 | G-A | HET | ||
| SBS | Chr11 | 1008275 | T-C | HET | ||
| SBS | Chr11 | 16401321 | C-T | HET | ||
| SBS | Chr11 | 3901315 | T-C | HET | ||
| SBS | Chr11 | 4272875 | C-T | HET | ||
| SBS | Chr11 | 4440000 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g08420 |
| SBS | Chr11 | 9658132 | G-T | HET | ||
| SBS | Chr12 | 6775202 | G-A | HET | ||
| SBS | Chr2 | 28514415 | G-A | HET | ||
| SBS | Chr3 | 3087048 | A-G | HET | ||
| SBS | Chr4 | 10292766 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g18610 |
| SBS | Chr4 | 33734701 | T-G | HET | ||
| SBS | Chr5 | 12918775 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g22770 |
| SBS | Chr5 | 9584201 | A-T | HET | ||
| SBS | Chr6 | 16716694 | G-A | Homo | STOP_GAINED | LOC_Os06g29280 |
| SBS | Chr6 | 26922513 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os06g44590 |
| SBS | Chr6 | 5918407 | G-C | HET | ||
| SBS | Chr6 | 6172782 | T-A | HET | ||
| SBS | Chr7 | 12542329 | T-A | HET | ||
| SBS | Chr7 | 18986813 | G-C | Homo | ||
| SBS | Chr7 | 7447494 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g12980 |
| SBS | Chr8 | 10779077 | G-A | HET | ||
| SBS | Chr8 | 2110557 | C-T | HET | ||
| SBS | Chr8 | 8681189 | C-T | HET | ||
| SBS | Chr9 | 19950493 | C-G | Homo |
Deletions: 14
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 471050 | 471055 | 5 | LOC_Os12g01770 |
| Deletion | Chr10 | 583828 | 583833 | 5 | LOC_Os10g01920 |
| Deletion | Chr3 | 3801001 | 3966000 | 164999 | 30 |
| Deletion | Chr12 | 4273678 | 4273691 | 13 | LOC_Os12g08410 |
| Deletion | Chr3 | 6107620 | 6107740 | 120 | |
| Deletion | Chr10 | 6257641 | 6257642 | 1 | |
| Deletion | Chr7 | 10071021 | 10071030 | 9 | LOC_Os07g17140 |
| Deletion | Chr1 | 10098199 | 10098202 | 3 | |
| Deletion | Chr11 | 10810001 | 10834000 | 23999 | 3 |
| Deletion | Chr4 | 18438001 | 18452000 | 13999 | 2 |
| Deletion | Chr7 | 20634374 | 20634390 | 16 | |
| Deletion | Chr7 | 27635069 | 27635070 | 1 | |
| Deletion | Chr5 | 29936290 | 29936291 | 1 | |
| Deletion | Chr1 | 30076812 | 30076820 | 8 | LOC_Os01g52330 |
Insertions: 11
Inversions: 12
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr5 | 2094385 | 2094497 | |
| Inversion | Chr1 | 2603026 | 2603346 | LOC_Os01g05500 |
| Inversion | Chr10 | 10588753 | 10589088 | LOC_Os10g20900 |
| Inversion | Chr5 | 11208019 | 11208180 | |
| Inversion | Chr5 | 13141975 | 13142136 | LOC_Os05g23050 |
| Inversion | Chr2 | 18772930 | 18773278 | LOC_Os02g31290 |
| Inversion | Chr8 | 20156748 | 20157150 | LOC_Os08g32540 |
| Inversion | Chr12 | 23808122 | 23808452 | LOC_Os12g38750 |
| Inversion | Chr11 | 27724639 | 27792555 | LOC_Os11g45820 |
| Inversion | Chr6 | 28757032 | 28757193 | LOC_Os06g47470 |
| Inversion | Chr2 | 29808827 | 29809132 | |
| Inversion | Chr1 | 31169785 | 31170249 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 916018 | Chr3 | 3015431 | LOC_Os11g02778 |
| Translocation | Chr9 | 7389228 | Chr2 | 13965460 | |
| Translocation | Chr12 | 18566076 | Chr5 | 10266652 | 2 |
