Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1745-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1745-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 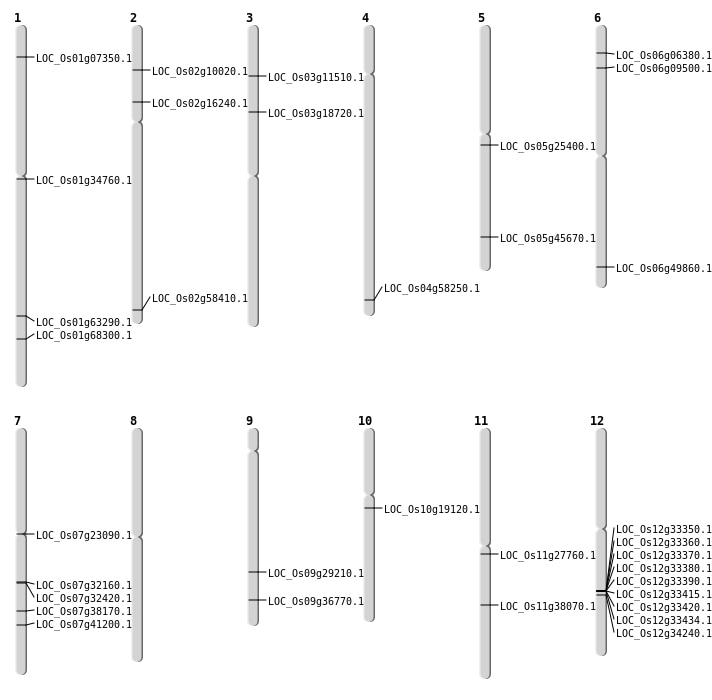
|
| Variant Information |
|---|
Single base substitutions: 36
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12513187 | G-A | Homo | ||
| SBS | Chr1 | 21428496 | C-G | HET | ||
| SBS | Chr1 | 25740756 | C-T | HET | ||
| SBS | Chr1 | 36428938 | G-A | Homo | ||
| SBS | Chr1 | 38121088 | T-A | Homo | ||
| SBS | Chr1 | 8673298 | C-T | Homo | ||
| SBS | Chr10 | 21315344 | G-A | HET | ||
| SBS | Chr10 | 4409768 | C-A | HET | ||
| SBS | Chr10 | 5793397 | T-C | HET | ||
| SBS | Chr10 | 9736295 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os10g19120 |
| SBS | Chr11 | 19215630 | G-A | HET | ||
| SBS | Chr11 | 24909768 | T-C | Homo | ||
| SBS | Chr12 | 13587308 | G-T | HET | ||
| SBS | Chr12 | 13587309 | A-T | HET | ||
| SBS | Chr12 | 1834684 | C-T | Homo | ||
| SBS | Chr12 | 20909684 | C-T | HET | ||
| SBS | Chr12 | 8813641 | A-G | HET | ||
| SBS | Chr3 | 10352975 | A-G | HET | ||
| SBS | Chr3 | 19808468 | C-T | HET | ||
| SBS | Chr3 | 28413274 | T-A | Homo | ||
| SBS | Chr4 | 17323104 | G-C | Homo | ||
| SBS | Chr4 | 2698658 | T-C | Homo | ||
| SBS | Chr4 | 3513493 | A-T | HET | ||
| SBS | Chr5 | 11296794 | G-A | Homo | ||
| SBS | Chr5 | 15305393 | G-A | Homo | ||
| SBS | Chr5 | 28988896 | G-A | HET | ||
| SBS | Chr6 | 21014105 | C-T | Homo | ||
| SBS | Chr6 | 28304037 | G-A | Homo | ||
| SBS | Chr6 | 30182936 | T-C | Homo | ||
| SBS | Chr6 | 8430281 | T-A | Homo | ||
| SBS | Chr7 | 13021238 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23090 |
| SBS | Chr7 | 25876419 | C-A | HET | ||
| SBS | Chr7 | 3961427 | A-T | HET | ||
| SBS | Chr8 | 10921252 | C-G | HET | ||
| SBS | Chr8 | 3005367 | A-G | Homo | ||
| SBS | Chr9 | 15237250 | C-T | Homo |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 2449093 | 2449094 | 1 | |
| Deletion | Chr12 | 8270209 | 8270213 | 4 | |
| Deletion | Chr3 | 16007766 | 16007768 | 2 | |
| Deletion | Chr2 | 17933877 | 17933882 | 5 | |
| Deletion | Chr12 | 20169001 | 20205000 | 35999 | 8 |
| Deletion | Chr6 | 26594184 | 26594188 | 4 | |
| Deletion | Chr5 | 29806517 | 29806519 | 2 | |
| Deletion | Chr6 | 30184219 | 30184228 | 9 | LOC_Os06g49860 |
| Deletion | Chr3 | 30461887 | 30461888 | 1 | |
| Deletion | Chr6 | 30506747 | 30506749 | 2 | |
| Deletion | Chr1 | 35180392 | 35180397 | 5 | |
| Deletion | Chr1 | 36689923 | 36689927 | 4 | LOC_Os01g63290 |
| Deletion | Chr1 | 41746810 | 41746812 | 2 |
Insertions: 5
Inversions: 32
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 1055292 | 1055560 | |
| Inversion | Chr3 | 2216203 | 2216421 | |
| Inversion | Chr6 | 2975611 | 2975744 | LOC_Os06g06380 |
| Inversion | Chr1 | 3468761 | 3469062 | LOC_Os01g07350 |
| Inversion | Chr7 | 3685202 | 3685394 | |
| Inversion | Chr6 | 4836222 | 4836677 | 2 |
| Inversion | Chr2 | 5209922 | 5210082 | LOC_Os02g10020 |
| Inversion | Chr3 | 5962253 | 5962363 | LOC_Os03g11510 |
| Inversion | Chr7 | 6088637 | 6088847 | |
| Inversion | Chr3 | 6774351 | 6774474 | |
| Inversion | Chr3 | 7052180 | 7052613 | |
| Inversion | Chr10 | 10462324 | 10467358 | |
| Inversion | Chr10 | 10462326 | 10467364 | |
| Inversion | Chr3 | 10478672 | 10479080 | LOC_Os03g18720 |
| Inversion | Chr11 | 15980361 | 15980464 | LOC_Os11g27760 |
| Inversion | Chr11 | 17086511 | 17086799 | |
| Inversion | Chr8 | 17859058 | 17859423 | |
| Inversion | Chr7 | 19098570 | 19098809 | LOC_Os07g32160 |
| Inversion | Chr7 | 19288663 | 19288937 | LOC_Os07g32420 |
| Inversion | Chr1 | 20275174 | 20275328 | |
| Inversion | Chr1 | 21195065 | 21195196 | |
| Inversion | Chr9 | 21222338 | 21222502 | LOC_Os09g36770 |
| Inversion | Chr7 | 22889184 | 22889333 | 2 |
| Inversion | Chr7 | 24666778 | 24667159 | LOC_Os07g41200 |
| Inversion | Chr5 | 26467526 | 26467758 | 2 |
| Inversion | Chr5 | 26663917 | 26664230 | |
| Inversion | Chr6 | 27614936 | 27615092 | |
| Inversion | Chr2 | 28800820 | 28852568 | |
| Inversion | Chr2 | 32553286 | 32553606 | |
| Inversion | Chr4 | 34684766 | 34685003 | LOC_Os04g58250 |
| Inversion | Chr2 | 35728740 | 35729184 | LOC_Os02g58410 |
| Inversion | Chr1 | 39683051 | 39683260 | LOC_Os01g68300 |
Translocations: 7
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 7337585 | Chr5 | 14772935 | 2 |
| Translocation | Chr12 | 8582247 | Chr1 | 19169623 | 2 |
| Translocation | Chr2 | 9236367 | Chr1 | 9625629 | LOC_Os02g16240 |
| Translocation | Chr7 | 16542927 | Chr2 | 28800811 | |
| Translocation | Chr12 | 20757990 | Chr9 | 17750447 | 2 |
| Translocation | Chr12 | 20759698 | Chr9 | 17750444 | 2 |
| Translocation | Chr11 | 22579323 | Chr8 | 3761938 | LOC_Os11g38070 |
