Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN1749-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN1749-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 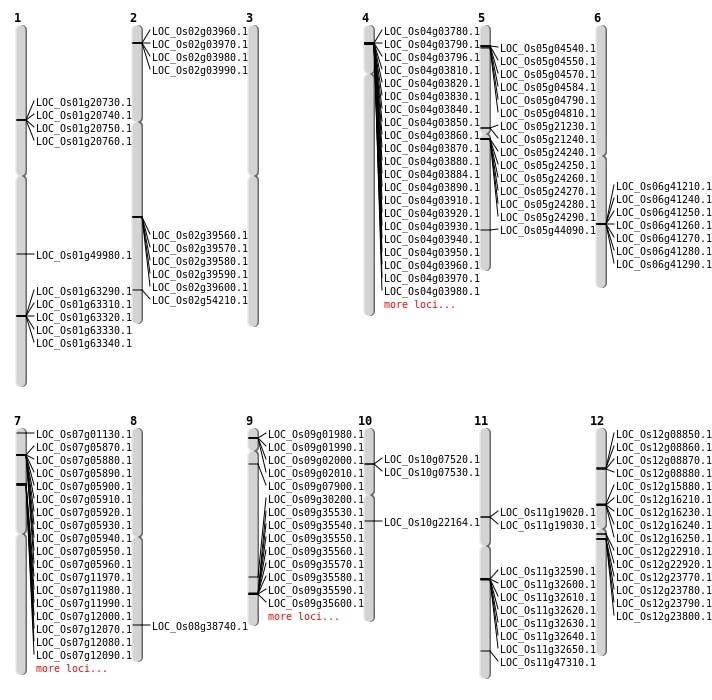
|
| Variant Information |
|---|
Single base substitutions: 34
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12403421 | C-T | Homo | ||
| SBS | Chr1 | 20297732 | G-A | Homo | ||
| SBS | Chr1 | 27999383 | A-G | HET | ||
| SBS | Chr1 | 28224398 | T-G | HET | ||
| SBS | Chr1 | 28558870 | C-A | HET | ||
| SBS | Chr1 | 28720416 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g49980 |
| SBS | Chr1 | 30834470 | A-T | HET | ||
| SBS | Chr10 | 2777775 | C-A | HET | ||
| SBS | Chr11 | 11360468 | T-G | Homo | ||
| SBS | Chr11 | 28452893 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os11g47310 |
| SBS | Chr12 | 14535197 | G-A | Homo | ||
| SBS | Chr12 | 16390686 | G-A | Homo | ||
| SBS | Chr12 | 16390687 | G-A | Homo | ||
| SBS | Chr2 | 15093320 | C-T | Homo | ||
| SBS | Chr2 | 18231471 | T-A | Homo | ||
| SBS | Chr2 | 18233546 | G-T | Homo | ||
| SBS | Chr3 | 34481605 | T-A | HET | ||
| SBS | Chr4 | 13030639 | T-A | HET | ||
| SBS | Chr4 | 19532859 | C-A | HET | ||
| SBS | Chr5 | 19614243 | T-A | HET | ||
| SBS | Chr5 | 25636699 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g44090 |
| SBS | Chr5 | 28615259 | G-A | HET | ||
| SBS | Chr6 | 14208582 | C-A | Homo | ||
| SBS | Chr6 | 16008067 | G-T | Homo | ||
| SBS | Chr6 | 3488633 | T-C | Homo | ||
| SBS | Chr6 | 4773429 | C-G | Homo | ||
| SBS | Chr7 | 14172978 | G-A | HET | ||
| SBS | Chr8 | 3469694 | A-T | HET | ||
| SBS | Chr9 | 11953749 | A-C | HET | ||
| SBS | Chr9 | 13644742 | C-A | HET | ||
| SBS | Chr9 | 19728883 | C-A | Homo | ||
| SBS | Chr9 | 4021510 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07900 |
| SBS | Chr9 | 5370254 | T-C | HET | ||
| SBS | Chr9 | 9795933 | A-T | HET |
Deletions: 37
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 683001 | 701000 | 17999 | 4 |
| Deletion | Chr4 | 1683001 | 2083000 | 399999 | 58 |
| Deletion | Chr2 | 1691001 | 1724000 | 32999 | 4 |
| Deletion | Chr5 | 2106001 | 2148000 | 41999 | 4 |
| Deletion | Chr1 | 2109201 | 2109202 | 1 | |
| Deletion | Chr5 | 2278001 | 2307000 | 28999 | 2 |
| Deletion | Chr7 | 2822001 | 2888000 | 65999 | 10 |
| Deletion | Chr10 | 4001001 | 4017000 | 15999 | 2 |
| Deletion | Chr12 | 4572001 | 4612000 | 39999 | 4 |
| Deletion | Chr4 | 4645001 | 4785000 | 139999 | 24 |
| Deletion | Chr7 | 6629001 | 6660000 | 30999 | 4 |
| Deletion | Chr7 | 6740001 | 6768000 | 27999 | 5 |
| Deletion | Chr1 | 7835878 | 7835906 | 28 | |
| Deletion | Chr7 | 7953256 | 7953260 | 4 | |
| Deletion | Chr12 | 9275001 | 9292000 | 16999 | 5 |
| Deletion | Chr11 | 10840001 | 10852000 | 11999 | 2 |
| Deletion | Chr11 | 11175126 | 11175128 | 2 | |
| Deletion | Chr10 | 11450689 | 11450692 | 3 | LOC_Os10g22164 |
| Deletion | Chr11 | 11519683 | 11519701 | 18 | |
| Deletion | Chr1 | 11530001 | 11578000 | 47999 | 4 |
| Deletion | Chr5 | 12555001 | 12581000 | 25999 | 2 |
| Deletion | Chr12 | 12933001 | 12957000 | 23999 | 2 |
| Deletion | Chr5 | 13336658 | 13336698 | 40 | |
| Deletion | Chr12 | 13505001 | 13531000 | 25999 | 4 |
| Deletion | Chr5 | 14023001 | 14051000 | 27999 | 6 |
| Deletion | Chr1 | 16567799 | 16567806 | 7 | |
| Deletion | Chr11 | 19231001 | 19285000 | 53999 | 7 |
| Deletion | Chr9 | 20407001 | 20497000 | 89999 | 10 |
| Deletion | Chr12 | 21394293 | 21394294 | 1 | |
| Deletion | Chr9 | 22472001 | 22501000 | 28999 | 3 |
| Deletion | Chr2 | 23880001 | 23903000 | 22999 | 5 |
| Deletion | Chr8 | 24497918 | 24497919 | 1 | LOC_Os08g38740 |
| Deletion | Chr6 | 24659001 | 24712000 | 52999 | 7 |
| Deletion | Chr8 | 24682897 | 24682903 | 6 | |
| Deletion | Chr4 | 25094406 | 25094412 | 6 | |
| Deletion | Chr4 | 30055130 | 30055131 | 1 | |
| Deletion | Chr1 | 36690001 | 36714000 | 23999 | 5 |
Insertions: 6
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 2594131 | 3292520 | |
| Inversion | Chr8 | 2594171 | 3292604 | |
| Inversion | Chr9 | 8301496 | 8301789 | |
| Inversion | Chr9 | 18381953 | 18382227 | LOC_Os09g30200 |
| Inversion | Chr9 | 21169670 | 21169857 | |
| Inversion | Chr11 | 24266602 | 24267019 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 77239 | Chr4 | 2430861 | LOC_Os07g01130 |
| Translocation | Chr7 | 77246 | Chr4 | 2430857 | LOC_Os07g01130 |
| Translocation | Chr12 | 9050771 | Chr2 | 33237740 | 2 |
| Translocation | Chr10 | 11150579 | Chr7 | 1494654 | |
| Translocation | Chr7 | 25192596 | Chr2 | 19585394 | LOC_Os07g42080 |
| Translocation | Chr12 | 25370427 | Chr8 | 15270537 |
