Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2082-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2082-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 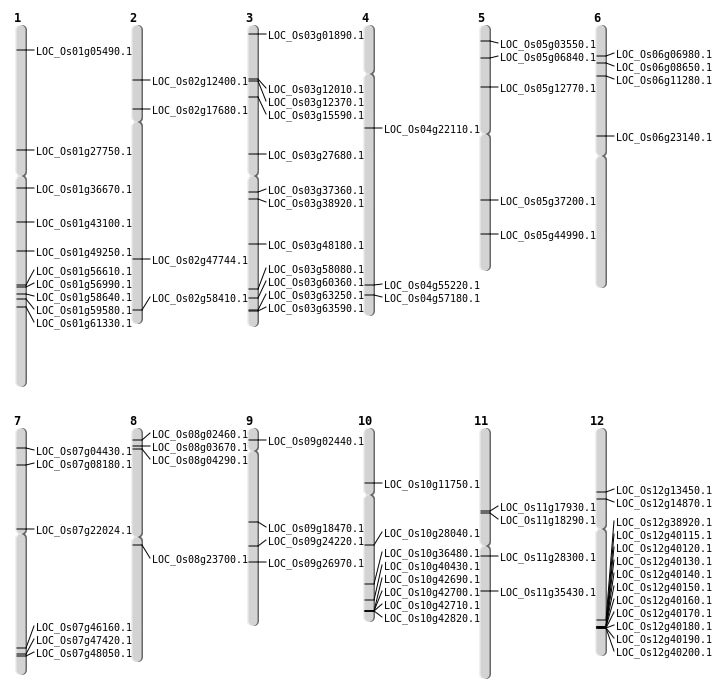
|
| Variant Information |
|---|
Single base substitutions: 38
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 17132835 | G-T | HET | ||
| SBS | Chr1 | 17377836 | G-A | HET | ||
| SBS | Chr1 | 3938259 | G-A | HET | ||
| SBS | Chr1 | 42279521 | C-T | HET | ||
| SBS | Chr10 | 5011897 | G-C | Homo | ||
| SBS | Chr11 | 10878140 | T-A | HET | ||
| SBS | Chr11 | 10878142 | T-A | HET | ||
| SBS | Chr11 | 14198643 | C-T | HET | ||
| SBS | Chr11 | 19301208 | G-A | HET | ||
| SBS | Chr11 | 5569346 | G-A | HET | ||
| SBS | Chr11 | 9850285 | G-A | HET | ||
| SBS | Chr11 | 9850286 | C-A | HET | ||
| SBS | Chr12 | 3482270 | G-A | Homo | ||
| SBS | Chr12 | 8514016 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g14870 |
| SBS | Chr2 | 18802266 | G-T | HET | ||
| SBS | Chr2 | 22411001 | C-T | HET | ||
| SBS | Chr2 | 25688594 | A-G | HET | ||
| SBS | Chr2 | 5279569 | A-T | HET | ||
| SBS | Chr2 | 6562667 | C-T | HET | ||
| SBS | Chr3 | 7615597 | C-T | HET | ||
| SBS | Chr4 | 29380342 | A-T | Homo | ||
| SBS | Chr5 | 11163452 | A-G | Homo | ||
| SBS | Chr5 | 15928649 | A-G | HET | ||
| SBS | Chr5 | 18363274 | A-T | Homo | ||
| SBS | Chr5 | 3584793 | A-G | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g06840 |
| SBS | Chr5 | 6362415 | G-T | HET | ||
| SBS | Chr6 | 16003297 | T-A | HET | ||
| SBS | Chr6 | 17722353 | G-A | HET | ||
| SBS | Chr6 | 19518741 | A-T | HET | ||
| SBS | Chr6 | 5913473 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g11280 |
| SBS | Chr7 | 11946329 | C-T | HET | ||
| SBS | Chr7 | 16637703 | G-T | HET | ||
| SBS | Chr7 | 8893194 | C-T | HET | ||
| SBS | Chr8 | 11518882 | C-G | Homo | ||
| SBS | Chr9 | 12349152 | C-T | HET | ||
| SBS | Chr9 | 1238754 | T-A | HET | ||
| SBS | Chr9 | 1672048 | T-A | HET | ||
| SBS | Chr9 | 3527943 | A-T | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 625030 | 625032 | 2 | |
| Deletion | Chr11 | 2523732 | 2523736 | 4 | |
| Deletion | Chr7 | 2950748 | 2950749 | 1 | |
| Deletion | Chr5 | 5452772 | 5452784 | 12 | |
| Deletion | Chr7 | 9156962 | 9156967 | 5 | |
| Deletion | Chr11 | 10304413 | 10304414 | 1 | LOC_Os11g18290 |
| Deletion | Chr6 | 13134493 | 13134494 | 1 | |
| Deletion | Chr8 | 19155815 | 19155819 | 4 | |
| Deletion | Chr10 | 23027001 | 23036000 | 8999 | 3 |
| Deletion | Chr12 | 24801001 | 24881000 | 79999 | 9 |
| Deletion | Chr1 | 40124780 | 40124787 | 7 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 22180356 | 22180357 | 2 | |
| Insertion | Chr8 | 3980778 | 3980778 | 1 | |
| Insertion | Chr9 | 12954539 | 12954569 | 31 |
Inversions: 122
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 49470 | 49815 | |
| Inversion | Chr6 | 166888 | 167036 | |
| Inversion | Chr3 | 552155 | 552329 | LOC_Os03g01890 |
| Inversion | Chr3 | 963105 | 963429 | |
| Inversion | Chr8 | 984849 | 985048 | LOC_Os08g02460 |
| Inversion | Chr9 | 1012229 | 1012487 | LOC_Os09g02440 |
| Inversion | Chr2 | 1101280 | 1101469 | |
| Inversion | Chr5 | 1501140 | 1501387 | LOC_Os05g03550 |
| Inversion | Chr8 | 1748106 | 1748262 | LOC_Os08g03670 |
| Inversion | Chr7 | 1964354 | 1964571 | LOC_Os07g04430 |
| Inversion | Chr8 | 2100081 | 2100351 | LOC_Os08g04290 |
| Inversion | Chr7 | 2185270 | 2185501 | |
| Inversion | Chr4 | 2460818 | 2461243 | |
| Inversion | Chr3 | 2537465 | 2537908 | |
| Inversion | Chr1 | 2591911 | 2592137 | LOC_Os01g05490 |
| Inversion | Chr4 | 3283468 | 3283614 | |
| Inversion | Chr6 | 3314924 | 3315084 | LOC_Os06g06980 |
| Inversion | Chr8 | 4056625 | 4056754 | |
| Inversion | Chr7 | 4158479 | 4158924 | LOC_Os07g08180 |
| Inversion | Chr6 | 4313972 | 4314125 | LOC_Os06g08650 |
| Inversion | Chr11 | 5095874 | 5472681 | |
| Inversion | Chr11 | 5095902 | 5472691 | |
| Inversion | Chr1 | 5120876 | 5120990 | |
| Inversion | Chr7 | 5207884 | 5208057 | |
| Inversion | Chr10 | 5276765 | 5276981 | |
| Inversion | Chr3 | 6292378 | 6292685 | LOC_Os03g12010 |
| Inversion | Chr2 | 6470032 | 6470541 | LOC_Os02g12400 |
| Inversion | Chr10 | 6509157 | 6509518 | LOC_Os10g11750 |
| Inversion | Chr3 | 6540815 | 6541046 | LOC_Os03g12370 |
| Inversion | Chr2 | 7082503 | 7082980 | |
| Inversion | Chr4 | 7177307 | 7177497 | |
| Inversion | Chr5 | 7326861 | 7327079 | LOC_Os05g12770 |
| Inversion | Chr9 | 7407215 | 7407525 | |
| Inversion | Chr12 | 7539872 | 7540133 | LOC_Os12g13450 |
| Inversion | Chr6 | 8091421 | 8091591 | |
| Inversion | Chr3 | 8597245 | 8597396 | 2 |
| Inversion | Chr8 | 8794399 | 9048637 | |
| Inversion | Chr8 | 8794661 | 9048643 | |
| Inversion | Chr11 | 8860276 | 8860478 | |
| Inversion | Chr9 | 9418983 | 9419191 | |
| Inversion | Chr11 | 10068383 | 10068641 | LOC_Os11g17930 |
| Inversion | Chr2 | 10183452 | 10184021 | LOC_Os02g17680 |
| Inversion | Chr9 | 11322747 | 11323066 | LOC_Os09g18470 |
| Inversion | Chr7 | 12322094 | 12322285 | LOC_Os07g22024 |
| Inversion | Chr8 | 12325631 | 12325796 | |
| Inversion | Chr4 | 12529775 | 12530285 | LOC_Os04g22110 |
| Inversion | Chr10 | 13445495 | 13445652 | |
| Inversion | Chr10 | 13445926 | 13446130 | |
| Inversion | Chr6 | 13498310 | 13498733 | 2 |
| Inversion | Chr8 | 14333724 | 14334050 | LOC_Os08g23700 |
| Inversion | Chr9 | 14385731 | 14385853 | LOC_Os09g24220 |
| Inversion | Chr11 | 14507601 | 14507976 | |
| Inversion | Chr10 | 14554336 | 14554503 | LOC_Os10g28040 |
| Inversion | Chr8 | 15452616 | 15452896 | |
| Inversion | Chr1 | 15482019 | 15482272 | LOC_Os01g27750 |
| Inversion | Chr9 | 15684396 | 15684638 | |
| Inversion | Chr3 | 15864324 | 15864515 | LOC_Os03g27680 |
| Inversion | Chr11 | 16278074 | 16278346 | LOC_Os11g28300 |
| Inversion | Chr9 | 16399172 | 16399501 | LOC_Os09g26970 |
| Inversion | Chr3 | 16659857 | 16660170 | |
| Inversion | Chr9 | 17118649 | 17118912 | |
| Inversion | Chr12 | 18016071 | 18016561 | |
| Inversion | Chr5 | 18374352 | 18374515 | |
| Inversion | Chr8 | 18652352 | 18652462 | |
| Inversion | Chr2 | 18664859 | 18665023 | |
| Inversion | Chr5 | 19496421 | 19496718 | LOC_Os05g33260 |
| Inversion | Chr10 | 19527548 | 19527680 | LOC_Os10g36480 |
| Inversion | Chr10 | 20288139 | 20288314 | |
| Inversion | Chr1 | 20358027 | 20358353 | LOC_Os01g36670 |
| Inversion | Chr2 | 20394617 | 20394783 | |
| Inversion | Chr4 | 20661864 | 20662177 | |
| Inversion | Chr3 | 20708488 | 20708811 | LOC_Os03g37360 |
| Inversion | Chr11 | 20762298 | 20762399 | LOC_Os11g35430 |
| Inversion | Chr7 | 21473924 | 21474084 | |
| Inversion | Chr3 | 21619985 | 21620201 | LOC_Os03g38920 |
| Inversion | Chr10 | 21658659 | 21658949 | LOC_Os10g40430 |
| Inversion | Chr5 | 21751672 | 21752101 | LOC_Os05g37200 |
| Inversion | Chr2 | 21773845 | 21774052 | |
| Inversion | Chr5 | 21912266 | 21912530 | |
| Inversion | Chr2 | 22261056 | 22261408 | |
| Inversion | Chr10 | 23090364 | 23090761 | LOC_Os10g42820 |
| Inversion | Chr12 | 23114732 | 23114935 | |
| Inversion | Chr11 | 23252627 | 23252788 | |
| Inversion | Chr3 | 23608246 | 23608438 | |
| Inversion | Chr12 | 23934068 | 24801673 | LOC_Os12g38920 |
| Inversion | Chr12 | 23934078 | 24883817 | 2 |
| Inversion | Chr5 | 24370682 | 24370859 | |
| Inversion | Chr1 | 24585922 | 24586058 | LOC_Os01g43100 |
| Inversion | Chr3 | 24642826 | 24643126 | |
| Inversion | Chr12 | 24882883 | 24883179 | LOC_Os12g40200 |
| Inversion | Chr1 | 25675320 | 25675665 | |
| Inversion | Chr6 | 25752705 | 25752847 | |
| Inversion | Chr8 | 25884745 | 25885031 | |
| Inversion | Chr5 | 26154276 | 26154527 | LOC_Os05g44990 |
| Inversion | Chr7 | 26245167 | 26245356 | |
| Inversion | Chr6 | 26601684 | 26601953 | |
| Inversion | Chr3 | 27418578 | 27418992 | LOC_Os03g48180 |
| Inversion | Chr7 | 27545916 | 27546312 | LOC_Os07g46160 |
| Inversion | Chr2 | 27733679 | 27733790 | |
| Inversion | Chr1 | 28314760 | 28314893 | LOC_Os01g49250 |
| Inversion | Chr7 | 28354724 | 28354883 | LOC_Os07g47420 |
| Inversion | Chr8 | 28383641 | 28384097 | |
| Inversion | Chr7 | 28689581 | 28689836 | LOC_Os07g48050 |
| Inversion | Chr2 | 28734129 | 28734293 | |
| Inversion | Chr2 | 28820512 | 28820649 | |
| Inversion | Chr6 | 28880863 | 28881049 | |
| Inversion | Chr2 | 29191736 | 29192147 | LOC_Os02g47744 |
| Inversion | Chr1 | 32656915 | 32657121 | LOC_Os01g56610 |
| Inversion | Chr4 | 32827009 | 32827241 | LOC_Os04g55220 |
| Inversion | Chr1 | 32930536 | 32930743 | LOC_Os01g56990 |
| Inversion | Chr3 | 33009203 | 33009348 | |
| Inversion | Chr3 | 33067798 | 33067941 | LOC_Os03g58080 |
| Inversion | Chr1 | 33904209 | 33904339 | LOC_Os01g58640 |
| Inversion | Chr4 | 34068276 | 34068452 | LOC_Os04g57180 |
| Inversion | Chr3 | 34329303 | 34329542 | LOC_Os03g60360 |
| Inversion | Chr4 | 34431836 | 34432075 | |
| Inversion | Chr1 | 34463627 | 34463865 | LOC_Os01g59580 |
| Inversion | Chr1 | 34669086 | 34669214 | |
| Inversion | Chr1 | 35479751 | 35480187 | 2 |
| Inversion | Chr2 | 35729520 | 35729724 | LOC_Os02g58410 |
| Inversion | Chr3 | 35736033 | 35736345 | 2 |
| Inversion | Chr3 | 35909505 | 35909702 | LOC_Os03g63590 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 20607133 | Chr3 | 24684655 | |
| Translocation | Chr6 | 22188579 | Chr5 | 11897074 | |
| Translocation | Chr11 | 27840603 | Chr8 | 12265045 |
