Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2084-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2084-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 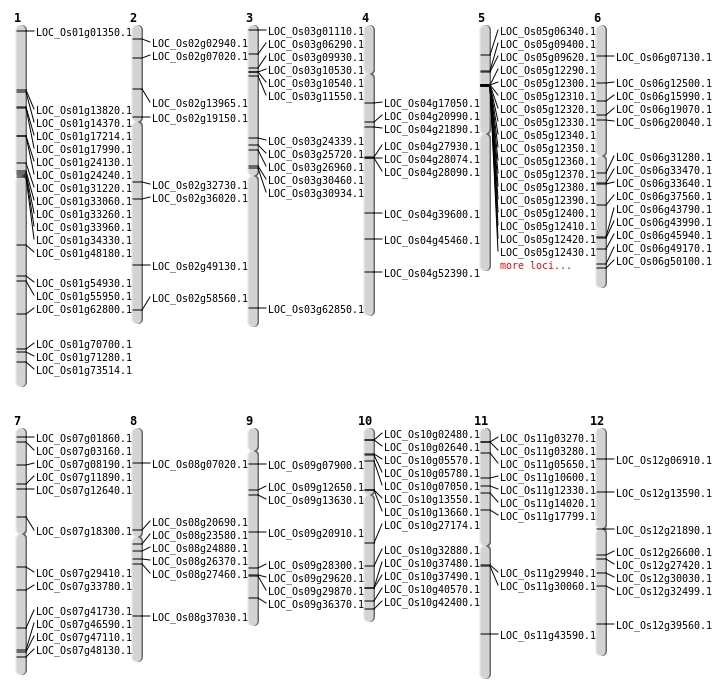
|
| Variant Information |
|---|
Single base substitutions: 26
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22693149 | G-A | HET | ||
| SBS | Chr1 | 23610257 | C-A | Homo | ||
| SBS | Chr1 | 26418162 | G-T | Homo | ||
| SBS | Chr1 | 34061950 | A-T | HET | ||
| SBS | Chr1 | 6796453 | C-T | HET | ||
| SBS | Chr10 | 16338348 | T-C | HET | ||
| SBS | Chr11 | 5813265 | G-A | Homo | ||
| SBS | Chr12 | 10403393 | C-A | HET | ||
| SBS | Chr12 | 16504527 | C-T | HET | ||
| SBS | Chr12 | 16993099 | T-C | HET | ||
| SBS | Chr12 | 19808277 | C-T | Homo | ||
| SBS | Chr3 | 17954274 | G-T | HET | ||
| SBS | Chr4 | 11813290 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g20990 |
| SBS | Chr4 | 15526854 | C-A | HET | ||
| SBS | Chr4 | 2730889 | C-A | HET | ||
| SBS | Chr5 | 13857679 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g24040 |
| SBS | Chr5 | 1678204 | A-T | HET | ||
| SBS | Chr5 | 20485892 | G-A | HET | ||
| SBS | Chr5 | 21856587 | A-T | HET | ||
| SBS | Chr7 | 2183347 | G-T | HET | ||
| SBS | Chr7 | 27830801 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g46590 |
| SBS | Chr7 | 29256247 | A-T | HET | ||
| SBS | Chr7 | 8540071 | A-G | HET | ||
| SBS | Chr8 | 18273947 | G-A | Homo | ||
| SBS | Chr8 | 27746761 | C-T | HET | ||
| SBS | Chr9 | 2208603 | T-C | Homo |
Deletions: 13
Insertions: 6
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr4 | 16490000 | 16490070 | 71 | LOC_Os04g27930 |
| Insertion | Chr4 | 22273373 | 22273429 | 57 | |
| Insertion | Chr6 | 13885049 | 13885056 | 8 | |
| Insertion | Chr7 | 27081879 | 27081880 | 2 | |
| Insertion | Chr8 | 5022341 | 5022343 | 3 | |
| Insertion | Chr9 | 6144438 | 6144438 | 1 |
Inversions: 228
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr3 | 75267 | 75601 | LOC_Os03g01110 |
| Inversion | Chr1 | 170756 | 170906 | LOC_Os01g01350 |
| Inversion | Chr7 | 519855 | 520020 | LOC_Os07g01860 |
| Inversion | Chr6 | 540694 | 540973 | |
| Inversion | Chr10 | 913563 | 913670 | LOC_Os10g02480 |
| Inversion | Chr10 | 1019583 | 1019704 | LOC_Os10g02640 |
| Inversion | Chr10 | 1072306 | 1072524 | |
| Inversion | Chr2 | 1151985 | 1152138 | LOC_Os02g02940 |
| Inversion | Chr5 | 1160526 | 1160841 | |
| Inversion | Chr3 | 1180858 | 1181051 | |
| Inversion | Chr7 | 1234745 | 1234926 | LOC_Os07g03160 |
| Inversion | Chr7 | 1775385 | 1775588 | |
| Inversion | Chr3 | 1828612 | 1828879 | |
| Inversion | Chr1 | 1873095 | 1873235 | |
| Inversion | Chr2 | 1926126 | 1926384 | |
| Inversion | Chr1 | 2081721 | 2081927 | |
| Inversion | Chr5 | 2151609 | 2151973 | |
| Inversion | Chr2 | 2199271 | 2199375 | |
| Inversion | Chr4 | 2475883 | 2476084 | |
| Inversion | Chr3 | 2485073 | 2485361 | |
| Inversion | Chr7 | 2492903 | 2493156 | |
| Inversion | Chr11 | 2566629 | 2566805 | LOC_Os11g05650 |
| Inversion | Chr10 | 2763849 | 2763999 | LOC_Os10g05570 |
| Inversion | Chr10 | 2913180 | 2913582 | LOC_Os10g05780 |
| Inversion | Chr3 | 3161818 | 3162023 | LOC_Os03g06290 |
| Inversion | Chr5 | 3170084 | 3170189 | |
| Inversion | Chr5 | 3197150 | 3197443 | |
| Inversion | Chr5 | 3245842 | 3246032 | LOC_Os05g06340 |
| Inversion | Chr12 | 3363995 | 3364131 | LOC_Os12g06910 |
| Inversion | Chr6 | 3416433 | 3416650 | LOC_Os06g07130 |
| Inversion | Chr2 | 3586428 | 3586890 | 2 |
| Inversion | Chr10 | 3702657 | 3702781 | LOC_Os10g07050 |
| Inversion | Chr8 | 3936537 | 3936659 | LOC_Os08g07020 |
| Inversion | Chr3 | 3966420 | 3966605 | |
| Inversion | Chr9 | 4026356 | 4026832 | LOC_Os09g07900 |
| Inversion | Chr9 | 4027739 | 4027903 | LOC_Os09g07900 |
| Inversion | Chr3 | 4068071 | 4068256 | |
| Inversion | Chr7 | 4181220 | 4181438 | LOC_Os07g08190 |
| Inversion | Chr7 | 4294242 | 4294597 | |
| Inversion | Chr7 | 4354237 | 4354392 | |
| Inversion | Chr9 | 4649693 | 4649986 | |
| Inversion | Chr7 | 4691947 | 4692235 | |
| Inversion | Chr12 | 4820248 | 4820503 | |
| Inversion | Chr3 | 4947254 | 4947563 | LOC_Os03g09930 |
| Inversion | Chr2 | 5003628 | 5003821 | |
| Inversion | Chr5 | 5034010 | 5034420 | |
| Inversion | Chr3 | 5115252 | 5115453 | |
| Inversion | Chr5 | 5276042 | 5276252 | LOC_Os05g09400 |
| Inversion | Chr5 | 5426509 | 5426699 | LOC_Os05g09620 |
| Inversion | Chr1 | 5457582 | 5457924 | |
| Inversion | Chr7 | 5542487 | 5542745 | |
| Inversion | Chr11 | 5814738 | 5815100 | LOC_Os11g10600 |
| Inversion | Chr4 | 6095422 | 6095544 | |
| Inversion | Chr4 | 6321713 | 6321966 | |
| Inversion | Chr1 | 6324990 | 6325182 | |
| Inversion | Chr3 | 6452510 | 6452829 | |
| Inversion | Chr7 | 6600583 | 6600879 | LOC_Os07g11890 |
| Inversion | Chr1 | 6705384 | 6705745 | |
| Inversion | Chr6 | 6788803 | 6789081 | 2 |
| Inversion | Chr11 | 6879143 | 6879361 | LOC_Os11g12330 |
| Inversion | Chr1 | 7047822 | 7048036 | |
| Inversion | Chr2 | 7157348 | 7157558 | |
| Inversion | Chr7 | 7230581 | 7230757 | LOC_Os07g12640 |
| Inversion | Chr9 | 7240508 | 7241080 | LOC_Os09g12650 |
| Inversion | Chr10 | 7335149 | 7335373 | LOC_Os10g13550 |
| Inversion | Chr10 | 7412234 | 7412717 | LOC_Os10g13660 |
| Inversion | Chr2 | 7487052 | 7487353 | |
| Inversion | Chr2 | 7620353 | 7620614 | LOC_Os02g13965 |
| Inversion | Chr12 | 7642397 | 7642524 | LOC_Os12g13590 |
| Inversion | Chr1 | 7753014 | 7753422 | LOC_Os01g13820 |
| Inversion | Chr11 | 7786280 | 7786495 | 2 |
| Inversion | Chr1 | 7918018 | 7918207 | |
| Inversion | Chr9 | 7929063 | 7929232 | LOC_Os09g13630 |
| Inversion | Chr1 | 8048573 | 8048843 | LOC_Os01g14370 |
| Inversion | Chr6 | 8053208 | 8053338 | |
| Inversion | Chr7 | 8454511 | 8454829 | |
| Inversion | Chr6 | 9094312 | 9094426 | LOC_Os06g15990 |
| Inversion | Chr11 | 9138001 | 9138349 | |
| Inversion | Chr4 | 9339510 | 9339688 | LOC_Os04g17050 |
| Inversion | Chr1 | 9891052 | 9891441 | LOC_Os01g17214 |
| Inversion | Chr11 | 9961616 | 9962126 | LOC_Os11g17799 |
| Inversion | Chr5 | 10123003 | 10123181 | LOC_Os05g17604 |
| Inversion | Chr6 | 10843934 | 10844416 | LOC_Os06g19070 |
| Inversion | Chr7 | 10845699 | 10845911 | LOC_Os07g18300 |
| Inversion | Chr5 | 10995457 | 10995772 | |
| Inversion | Chr2 | 11161876 | 11162357 | 2 |
| Inversion | Chr10 | 11477558 | 11477947 | |
| Inversion | Chr6 | 11492193 | 11492473 | LOC_Os06g20040 |
| Inversion | Chr3 | 11538505 | 11538620 | |
| Inversion | Chr12 | 11839641 | 11839911 | |
| Inversion | Chr4 | 11927436 | 11927681 | |
| Inversion | Chr2 | 12193081 | 12193291 | |
| Inversion | Chr1 | 12201286 | 12201440 | |
| Inversion | Chr12 | 12321597 | 12321797 | 2 |
| Inversion | Chr4 | 12397741 | 12398126 | LOC_Os04g21890 |
| Inversion | Chr8 | 12434349 | 12434721 | LOC_Os08g20690 |
| Inversion | Chr1 | 12506520 | 12506651 | |
| Inversion | Chr9 | 12585733 | 12586049 | LOC_Os09g20910 |
| Inversion | Chr3 | 12829325 | 12829455 | |
| Inversion | Chr1 | 12854793 | 12855005 | |
| Inversion | Chr9 | 12881994 | 12882209 | |
| Inversion | Chr2 | 13240373 | 13240531 | |
| Inversion | Chr12 | 13349248 | 13349390 | |
| Inversion | Chr12 | 13373077 | 13403852 | |
| Inversion | Chr9 | 13410649 | 13410882 | |
| Inversion | Chr1 | 13601380 | 13601724 | 2 |
| Inversion | Chr1 | 13664632 | 13664999 | LOC_Os01g24240 |
| Inversion | Chr3 | 13841498 | 13841680 | LOC_Os03g24339 |
| Inversion | Chr1 | 13936153 | 13936476 | |
| Inversion | Chr8 | 14273968 | 14274099 | LOC_Os08g23580 |
| Inversion | Chr10 | 14324621 | 14324764 | LOC_Os10g27174 |
| Inversion | Chr3 | 14720899 | 14721238 | 2 |
| Inversion | Chr9 | 14935930 | 14936100 | |
| Inversion | Chr8 | 15061827 | 15062109 | LOC_Os08g24880 |
| Inversion | Chr3 | 15414765 | 15415060 | LOC_Os03g26960 |
| Inversion | Chr2 | 15888593 | 15888923 | |
| Inversion | Chr8 | 16072816 | 16073074 | 2 |
| Inversion | Chr12 | 16139878 | 16140088 | LOC_Os12g27420 |
| Inversion | Chr5 | 16155063 | 16155310 | |
| Inversion | Chr11 | 16216861 | 16217181 | |
| Inversion | Chr2 | 16224803 | 16225157 | |
| Inversion | Chr4 | 16572078 | 16572305 | LOC_Os04g28074 |
| Inversion | Chr8 | 16765875 | 16766107 | LOC_Os08g27460 |
| Inversion | Chr1 | 17070781 | 17071067 | LOC_Os01g31220 |
| Inversion | Chr9 | 17150630 | 17151063 | |
| Inversion | Chr9 | 17186957 | 17187208 | LOC_Os09g28300 |
| Inversion | Chr10 | 17203666 | 17203935 | 2 |
| Inversion | Chr7 | 17268371 | 17268692 | 2 |
| Inversion | Chr5 | 17340654 | 17340901 | LOC_Os05g29974 |
| Inversion | Chr3 | 17380565 | 17380713 | LOC_Os03g30460 |
| Inversion | Chr1 | 17443633 | 17443969 | |
| Inversion | Chr11 | 17488824 | 17489292 | LOC_Os11g30060 |
| Inversion | Chr4 | 17558726 | 17558845 | |
| Inversion | Chr10 | 17610608 | 17610892 | LOC_Os10g33440 |
| Inversion | Chr3 | 17618776 | 17619092 | LOC_Os03g30934 |
| Inversion | Chr8 | 17642417 | 17642876 | |
| Inversion | Chr11 | 17849011 | 17849213 | |
| Inversion | Chr12 | 17965619 | 17966079 | LOC_Os12g30030 |
| Inversion | Chr9 | 18012354 | 18012539 | LOC_Os09g29620 |
| Inversion | Chr9 | 18164207 | 18164410 | LOC_Os09g29870 |
| Inversion | Chr1 | 18166023 | 18166184 | LOC_Os01g33060 |
| Inversion | Chr6 | 18202566 | 18202724 | LOC_Os06g31280 |
| Inversion | Chr1 | 18336182 | 18336524 | LOC_Os01g33260 |
| Inversion | Chr10 | 18508212 | 18508393 | |
| Inversion | Chr10 | 18765854 | 18766140 | |
| Inversion | Chr1 | 18782594 | 18782937 | |
| Inversion | Chr5 | 18786524 | 18786698 | LOC_Os05g32210 |
| Inversion | Chr9 | 18810997 | 18811343 | |
| Inversion | Chr1 | 18931302 | 18931489 | LOC_Os01g34330 |
| Inversion | Chr2 | 19441350 | 19441571 | LOC_Os02g32730 |
| Inversion | Chr6 | 19501800 | 19502039 | LOC_Os06g33470 |
| Inversion | Chr6 | 19585027 | 19585174 | LOC_Os06g33640 |
| Inversion | Chr12 | 19616724 | 19616928 | LOC_Os12g32499 |
| Inversion | Chr4 | 19833034 | 19833310 | |
| Inversion | Chr6 | 19997550 | 19997774 | |
| Inversion | Chr10 | 20070394 | 20070931 | 2 |
| Inversion | Chr10 | 20070996 | 20071150 | LOC_Os10g37490 |
| Inversion | Chr10 | 20071268 | 20071377 | LOC_Os10g37490 |
| Inversion | Chr7 | 20213961 | 20214098 | 2 |
| Inversion | Chr12 | 20468629 | 20468730 | |
| Inversion | Chr10 | 20892565 | 20892724 | |
| Inversion | Chr11 | 20968927 | 20969327 | |
| Inversion | Chr9 | 20990704 | 20990852 | LOC_Os09g36370 |
| Inversion | Chr9 | 21621411 | 21621607 | |
| Inversion | Chr2 | 21639074 | 21639234 | LOC_Os02g36020 |
| Inversion | Chr11 | 21713031 | 21713255 | |
| Inversion | Chr10 | 21725676 | 21725871 | LOC_Os10g40570 |
| Inversion | Chr6 | 22250569 | 22250700 | LOC_Os06g37560 |
| Inversion | Chr4 | 22347475 | 22347749 | |
| Inversion | Chr1 | 22818519 | 22818705 | |
| Inversion | Chr10 | 22820436 | 22820759 | LOC_Os10g42400 |
| Inversion | Chr12 | 23039652 | 23040115 | |
| Inversion | Chr7 | 23149650 | 23149810 | |
| Inversion | Chr1 | 23366201 | 23366469 | |
| Inversion | Chr7 | 23405285 | 23405417 | |
| Inversion | Chr8 | 23409591 | 23409748 | LOC_Os08g37030 |
| Inversion | Chr2 | 23589724 | 23589840 | |
| Inversion | Chr4 | 23599329 | 23599751 | 2 |
| Inversion | Chr5 | 23738804 | 23739035 | |
| Inversion | Chr1 | 23910011 | 23910199 | |
| Inversion | Chr7 | 23938062 | 23938232 | |
| Inversion | Chr5 | 24100066 | 24100311 | |
| Inversion | Chr6 | 24130481 | 24130649 | |
| Inversion | Chr12 | 24398007 | 24398206 | LOC_Os12g39560 |
| Inversion | Chr8 | 24852395 | 24852613 | |
| Inversion | Chr1 | 24884783 | 24884951 | |
| Inversion | Chr1 | 24962982 | 24963083 | |
| Inversion | Chr7 | 25008100 | 25008270 | LOC_Os07g41730 |
| Inversion | Chr11 | 25301588 | 25302165 | |
| Inversion | Chr11 | 25301590 | 25302034 | |
| Inversion | Chr11 | 25301793 | 25302473 | |
| Inversion | Chr12 | 26069404 | 26069603 | |
| Inversion | Chr11 | 26305315 | 26305750 | LOC_Os11g43590 |
| Inversion | Chr11 | 26305483 | 26305714 | LOC_Os11g43590 |
| Inversion | Chr6 | 26363239 | 26363642 | LOC_Os06g43790 |
| Inversion | Chr6 | 26383192 | 26383448 | |
| Inversion | Chr6 | 26497861 | 26498012 | LOC_Os06g43990 |
| Inversion | Chr6 | 26513341 | 26513576 | |
| Inversion | Chr4 | 26903583 | 26904094 | LOC_Os04g45460 |
| Inversion | Chr11 | 26958684 | 26958801 | |
| Inversion | Chr4 | 26997435 | 26997793 | |
| Inversion | Chr5 | 27040836 | 27040988 | |
| Inversion | Chr8 | 27402749 | 27402919 | |
| Inversion | Chr1 | 27593691 | 27593809 | LOC_Os01g48180 |
| Inversion | Chr6 | 27824424 | 27824715 | LOC_Os06g45940 |
| Inversion | Chr7 | 28174675 | 28174897 | LOC_Os07g47110 |
| Inversion | Chr2 | 28451286 | 28451391 | |
| Inversion | Chr7 | 28742712 | 28742827 | LOC_Os07g48130 |
| Inversion | Chr6 | 29793459 | 29793664 | LOC_Os06g49170 |
| Inversion | Chr2 | 30046623 | 30047045 | LOC_Os02g49130 |
| Inversion | Chr1 | 30090773 | 30091038 | |
| Inversion | Chr6 | 30348596 | 30348732 | LOC_Os06g50100 |
| Inversion | Chr4 | 31130483 | 31130722 | LOC_Os04g52390 |
| Inversion | Chr1 | 31593843 | 31594096 | LOC_Os01g54930 |
| Inversion | Chr1 | 32226610 | 32226888 | 2 |
| Inversion | Chr4 | 33159578 | 33159765 | |
| Inversion | Chr2 | 33456071 | 33456370 | |
| Inversion | Chr1 | 34394808 | 34395037 | |
| Inversion | Chr3 | 34428936 | 34429292 | |
| Inversion | Chr1 | 35180443 | 35180676 | |
| Inversion | Chr3 | 35555627 | 35555873 | LOC_Os03g62850 |
| Inversion | Chr2 | 35803806 | 35804031 | LOC_Os02g58560 |
| Inversion | Chr1 | 36370004 | 36370111 | LOC_Os01g62800 |
| Inversion | Chr1 | 37960801 | 37961102 | |
| Inversion | Chr1 | 38368093 | 38368422 | |
| Inversion | Chr1 | 40929061 | 40929441 | LOC_Os01g70700 |
| Inversion | Chr1 | 41249138 | 41249466 | LOC_Os01g71280 |
| Inversion | Chr1 | 42593825 | 43159146 | LOC_Os01g73514 |
Translocations: 12
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr4 | 3224461 | Chr3 | 5986400 | 2 |
| Translocation | Chr4 | 3224464 | Chr3 | 5987046 | 2 |
| Translocation | Chr6 | 3477642 | Chr1 | 10062145 | 2 |
| Translocation | Chr9 | 5040373 | Chr4 | 1632055 | |
| Translocation | Chr8 | 5818464 | Chr1 | 18686916 | 2 |
| Translocation | Chr11 | 14753919 | Chr6 | 9352430 | |
| Translocation | Chr12 | 15578263 | Chr1 | 42594682 | 2 |
| Translocation | Chr5 | 15913883 | Chr2 | 15512796 | |
| Translocation | Chr4 | 16584441 | Chr2 | 20382605 | LOC_Os04g28090 |
| Translocation | Chr11 | 17401312 | Chr2 | 4760397 | LOC_Os11g29940 |
| Translocation | Chr11 | 24408147 | Chr8 | 16614773 | |
| Translocation | Chr11 | 24439297 | Chr5 | 21302397 |
