Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2122-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2122-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 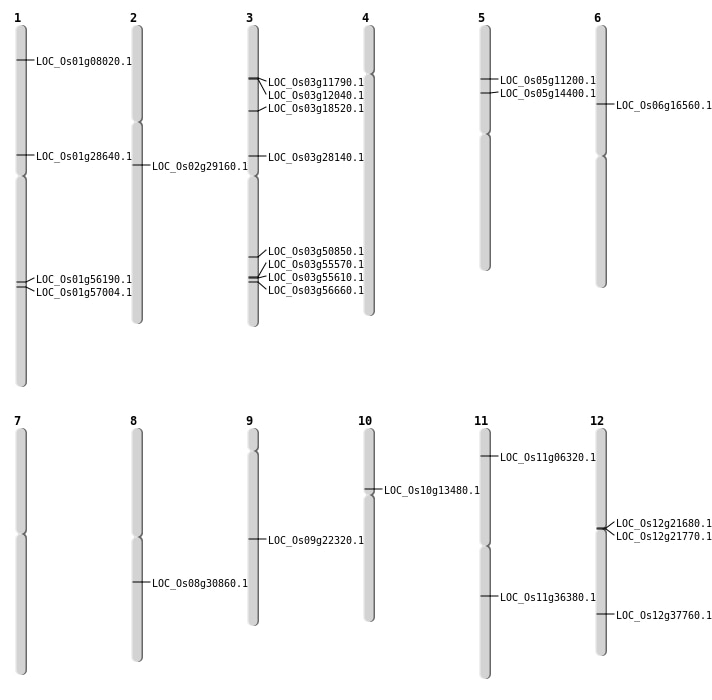
|
| Variant Information |
|---|
Single base substitutions: 33
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14634354 | T-C | Homo | ||
| SBS | Chr1 | 1905427 | C-A | HET | ||
| SBS | Chr1 | 32934485 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os01g57004 |
| SBS | Chr1 | 40505036 | C-A | HET | ||
| SBS | Chr10 | 22225039 | T-C | HET | ||
| SBS | Chr10 | 7283514 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g13480 |
| SBS | Chr11 | 21436071 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os11g36380 |
| SBS | Chr11 | 7515183 | G-A | Homo | ||
| SBS | Chr11 | 8796402 | G-T | Homo | ||
| SBS | Chr11 | 8796403 | G-T | Homo | ||
| SBS | Chr12 | 12180926 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21680 |
| SBS | Chr12 | 12248967 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g21770 |
| SBS | Chr12 | 4011765 | C-T | HET | ||
| SBS | Chr2 | 31961111 | G-A | HET | ||
| SBS | Chr2 | 8839659 | C-T | HET | ||
| SBS | Chr3 | 17337623 | G-A | HET | ||
| SBS | Chr3 | 19411728 | A-T | HET | ||
| SBS | Chr3 | 22478851 | G-A | HET | ||
| SBS | Chr4 | 11218635 | G-T | HET | ||
| SBS | Chr4 | 11787023 | G-T | Homo | ||
| SBS | Chr4 | 17674414 | G-T | HET | ||
| SBS | Chr4 | 17674415 | G-T | HET | ||
| SBS | Chr4 | 19107504 | C-T | HET | ||
| SBS | Chr4 | 29026301 | G-T | Homo | ||
| SBS | Chr4 | 29998927 | C-T | Homo | ||
| SBS | Chr5 | 14881394 | T-G | Homo | ||
| SBS | Chr5 | 3149560 | T-G | Homo | ||
| SBS | Chr6 | 29848564 | G-A | Homo | ||
| SBS | Chr6 | 8700362 | G-T | HET | ||
| SBS | Chr7 | 844457 | G-C | HET | ||
| SBS | Chr8 | 14317904 | G-A | HET | ||
| SBS | Chr9 | 22551727 | G-A | HET | ||
| SBS | Chr9 | 9383424 | C-T | Homo |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 3030735 | 3030742 | 7 | LOC_Os11g06320 |
| Deletion | Chr11 | 5148878 | 5148882 | 4 | |
| Deletion | Chr12 | 5253131 | 5253136 | 5 | |
| Deletion | Chr9 | 8048177 | 8048178 | 1 | |
| Deletion | Chr5 | 8115307 | 8115308 | 1 | LOC_Os05g14400 |
| Deletion | Chr11 | 8171348 | 8171351 | 3 | |
| Deletion | Chr5 | 8185683 | 8185684 | 1 | |
| Deletion | Chr11 | 8524919 | 8524923 | 4 | |
| Deletion | Chr10 | 9332673 | 9332675 | 2 | |
| Deletion | Chr8 | 10747831 | 10747940 | 109 | |
| Deletion | Chr10 | 13080787 | 13080794 | 7 | |
| Deletion | Chr9 | 13488195 | 13488200 | 5 | LOC_Os09g22320 |
| Deletion | Chr1 | 16895476 | 16895487 | 11 | |
| Deletion | Chr3 | 20954102 | 20954104 | 2 | |
| Deletion | Chr5 | 22795066 | 22795067 | 1 | |
| Deletion | Chr2 | 31187972 | 31187974 | 2 | |
| Deletion | Chr4 | 32989011 | 32989013 | 2 | |
| Deletion | Chr1 | 42179185 | 42179186 | 1 |
Insertions: 8
Inversions: 35
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 1131981 | 1132369 | |
| Inversion | Chr5 | 2229582 | 2229906 | |
| Inversion | Chr1 | 3898924 | 3899087 | LOC_Os01g08020 |
| Inversion | Chr11 | 4474964 | 4475333 | |
| Inversion | Chr3 | 6171010 | 6171254 | LOC_Os03g11790 |
| Inversion | Chr3 | 6318618 | 6318800 | 2 |
| Inversion | Chr5 | 6329999 | 6330243 | LOC_Os05g11200 |
| Inversion | Chr4 | 6910379 | 6910569 | |
| Inversion | Chr2 | 7074335 | 7074533 | |
| Inversion | Chr3 | 7555382 | 7555806 | |
| Inversion | Chr6 | 9499644 | 9499897 | LOC_Os06g16560 |
| Inversion | Chr7 | 10253252 | 10253542 | |
| Inversion | Chr3 | 10378654 | 10379112 | LOC_Os03g18520 |
| Inversion | Chr4 | 12054729 | 12055057 | |
| Inversion | Chr7 | 13474685 | 13475113 | |
| Inversion | Chr9 | 13820096 | 13820240 | |
| Inversion | Chr5 | 15725799 | 15726248 | |
| Inversion | Chr1 | 16025384 | 16025523 | LOC_Os01g28640 |
| Inversion | Chr3 | 16185771 | 16186277 | LOC_Os03g28140 |
| Inversion | Chr2 | 16238200 | 16238421 | |
| Inversion | Chr2 | 17276854 | 17277386 | 2 |
| Inversion | Chr8 | 19048744 | 19049237 | 2 |
| Inversion | Chr5 | 20795228 | 20795434 | |
| Inversion | Chr8 | 20882044 | 20882289 | |
| Inversion | Chr8 | 20882363 | 20882506 | |
| Inversion | Chr1 | 21661254 | 21661410 | |
| Inversion | Chr4 | 22990557 | 22990947 | |
| Inversion | Chr6 | 27432810 | 27433103 | |
| Inversion | Chr3 | 29045512 | 29045933 | LOC_Os03g50850 |
| Inversion | Chr4 | 30214981 | 31167600 | |
| Inversion | Chr4 | 30215004 | 31167603 | |
| Inversion | Chr3 | 31625994 | 31626279 | LOC_Os03g55570 |
| Inversion | Chr3 | 31662641 | 31662773 | LOC_Os03g55610 |
| Inversion | Chr3 | 32270918 | 32271099 | LOC_Os03g56660 |
| Inversion | Chr1 | 32351382 | 32351719 | LOC_Os01g56190 |
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 23183621 | Chr6 | 23504877 | LOC_Os12g37760 |
| Translocation | Chr5 | 27683386 | Chr3 | 10461181 |
