Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2124-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2124-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 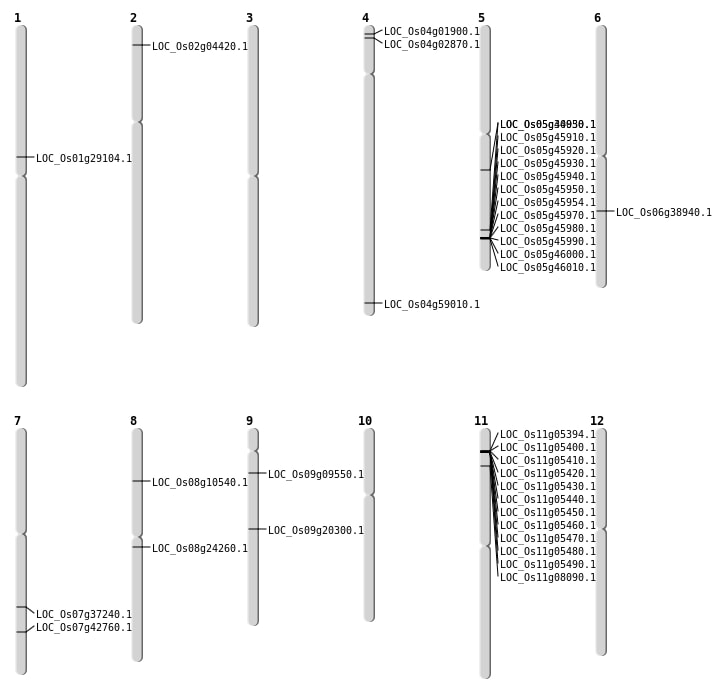
|
| Variant Information |
|---|
Single base substitutions: 50
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14647740 | C-A | Homo | ||
| SBS | Chr1 | 27111775 | G-C | Homo | ||
| SBS | Chr1 | 32373485 | C-T | HET | ||
| SBS | Chr1 | 42233505 | A-G | HET | ||
| SBS | Chr1 | 42250379 | A-G | HET | ||
| SBS | Chr10 | 15281431 | C-T | HET | ||
| SBS | Chr10 | 1553072 | C-G | HET | ||
| SBS | Chr10 | 1895170 | G-A | HET | ||
| SBS | Chr11 | 13854698 | T-C | HET | ||
| SBS | Chr11 | 18667803 | C-G | Homo | ||
| SBS | Chr12 | 10043662 | G-T | HET | ||
| SBS | Chr12 | 10729904 | T-C | HET | ||
| SBS | Chr12 | 13172279 | G-A | HET | ||
| SBS | Chr12 | 13580922 | A-G | HET | ||
| SBS | Chr12 | 14600119 | A-G | HET | ||
| SBS | Chr12 | 25418010 | A-G | HET | ||
| SBS | Chr12 | 27202446 | T-C | HET | ||
| SBS | Chr2 | 23833839 | G-A | HET | ||
| SBS | Chr3 | 1452384 | C-T | Homo | ||
| SBS | Chr3 | 1452385 | C-T | Homo | ||
| SBS | Chr3 | 17223868 | C-T | HET | ||
| SBS | Chr3 | 32082567 | C-T | Homo | ||
| SBS | Chr3 | 33204073 | T-A | Homo | ||
| SBS | Chr4 | 1121403 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g02870 |
| SBS | Chr4 | 1151375 | G-T | HET | ||
| SBS | Chr4 | 17030747 | C-A | HET | ||
| SBS | Chr4 | 25881095 | C-T | HET | ||
| SBS | Chr4 | 33358305 | T-C | HET | ||
| SBS | Chr4 | 566632 | G-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g01900 |
| SBS | Chr5 | 4354544 | C-G | Homo | ||
| SBS | Chr5 | 4794471 | C-T | Homo | ||
| SBS | Chr6 | 1475549 | A-T | HET | ||
| SBS | Chr6 | 15807072 | C-T | HET | ||
| SBS | Chr6 | 24180715 | C-T | Homo | ||
| SBS | Chr6 | 24796043 | C-T | HET | ||
| SBS | Chr6 | 3404481 | A-G | Homo | ||
| SBS | Chr6 | 7361456 | C-T | Homo | ||
| SBS | Chr7 | 22317659 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os07g37240 |
| SBS | Chr7 | 22710315 | G-A | HET | ||
| SBS | Chr7 | 28169912 | G-A | Homo | ||
| SBS | Chr8 | 10897774 | G-C | HET | ||
| SBS | Chr8 | 13468899 | A-T | Homo | ||
| SBS | Chr8 | 21896232 | G-A | HET | ||
| SBS | Chr8 | 21896233 | C-T | HET | ||
| SBS | Chr8 | 2542860 | T-A | HET | ||
| SBS | Chr8 | 6273689 | T-A | HET | ||
| SBS | Chr8 | 7516616 | T-C | Homo | ||
| SBS | Chr8 | 7729375 | C-T | HET | ||
| SBS | Chr9 | 10150671 | T-A | HET | ||
| SBS | Chr9 | 12180266 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g20300 |
Deletions: 13
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 1284473 | 1284474 | 1 | |
| Deletion | Chr11 | 2418001 | 2478000 | 59999 | 11 |
| Deletion | Chr6 | 3392164 | 3392171 | 7 | |
| Deletion | Chr9 | 5157407 | 5157408 | 1 | LOC_Os09g09550 |
| Deletion | Chr10 | 15989649 | 15989663 | 14 | |
| Deletion | Chr6 | 16185368 | 16185371 | 3 | |
| Deletion | Chr10 | 16786358 | 16786360 | 2 | |
| Deletion | Chr7 | 21615815 | 21615816 | 1 | |
| Deletion | Chr5 | 25522542 | 25522554 | 12 | |
| Deletion | Chr5 | 26605001 | 26675000 | 69999 | 11 |
| Deletion | Chr8 | 27002081 | 27002082 | 1 | |
| Deletion | Chr3 | 33655147 | 33655148 | 1 | |
| Deletion | Chr1 | 42199934 | 42199937 | 3 |
Insertions: 8
Inversions: 18
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 751228 | 751563 | |
| Inversion | Chr6 | 2326951 | 2327420 | |
| Inversion | Chr11 | 4234364 | 4234906 | LOC_Os11g08090 |
| Inversion | Chr8 | 6190538 | 6841074 | LOC_Os08g10540 |
| Inversion | Chr1 | 16298626 | 16299105 | LOC_Os01g29104 |
| Inversion | Chr5 | 17968672 | 17968828 | LOC_Os05g30950 |
| Inversion | Chr10 | 21838024 | 21838181 | |
| Inversion | Chr4 | 21868184 | 21868298 | |
| Inversion | Chr12 | 22454576 | 22454759 | |
| Inversion | Chr6 | 23099396 | 23099751 | LOC_Os06g38940 |
| Inversion | Chr6 | 23337495 | 23337876 | |
| Inversion | Chr7 | 25601066 | 25601434 | |
| Inversion | Chr7 | 25615795 | 25616035 | LOC_Os07g42760 |
| Inversion | Chr5 | 25616184 | 25616503 | LOC_Os05g44030 |
| Inversion | Chr3 | 27369389 | 27369652 | |
| Inversion | Chr5 | 28829664 | 28829783 | |
| Inversion | Chr1 | 34521012 | 34521160 | |
| Inversion | Chr4 | 35106854 | 35107118 | LOC_Os04g59010 |
Translocations: 3
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 14645205 | Chr4 | 3954961 | LOC_Os08g24260 |
| Translocation | Chr10 | 17670230 | Chr2 | 24459329 | |
| Translocation | Chr8 | 24363151 | Chr2 | 1947526 | 2 |
