Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2134-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2134-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 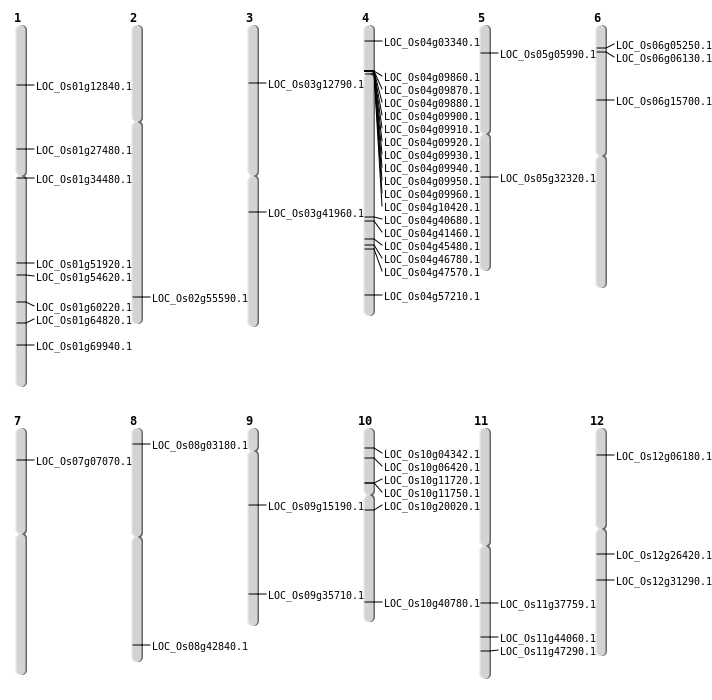
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13040605 | T-C | HET | ||
| SBS | Chr1 | 13452746 | G-A | HET | ||
| SBS | Chr1 | 17362919 | T-A | HET | ||
| SBS | Chr1 | 27031193 | G-A | HET | ||
| SBS | Chr1 | 34824219 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g60220 |
| SBS | Chr1 | 40660723 | C-T | Homo | ||
| SBS | Chr1 | 611375 | C-A | HET | ||
| SBS | Chr1 | 9721536 | G-A | HET | ||
| SBS | Chr10 | 10043080 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g20020 |
| SBS | Chr10 | 10646547 | A-T | HET | ||
| SBS | Chr10 | 10721180 | A-G | HET | ||
| SBS | Chr10 | 20005793 | T-A | HET | ||
| SBS | Chr10 | 3299296 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g06420 |
| SBS | Chr10 | 3798084 | C-G | HET | ||
| SBS | Chr10 | 4763696 | A-C | HET | ||
| SBS | Chr11 | 13365779 | C-T | HET | ||
| SBS | Chr11 | 14093859 | C-G | HET | ||
| SBS | Chr11 | 16062778 | A-T | HET | ||
| SBS | Chr11 | 24330815 | C-A | Homo | ||
| SBS | Chr11 | 26629087 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g44060 |
| SBS | Chr12 | 15442902 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g26420 |
| SBS | Chr12 | 19318396 | T-A | HET | ||
| SBS | Chr12 | 3614274 | A-G | Homo | ||
| SBS | Chr2 | 19706535 | C-T | HET | ||
| SBS | Chr2 | 21666373 | T-C | HET | ||
| SBS | Chr2 | 29404864 | G-A | HET | ||
| SBS | Chr2 | 30647501 | T-A | HET | ||
| SBS | Chr2 | 32614496 | T-C | HET | ||
| SBS | Chr3 | 23941125 | C-A | HET | ||
| SBS | Chr3 | 25141790 | C-T | HET | ||
| SBS | Chr3 | 31661015 | C-A | Homo | ||
| SBS | Chr3 | 4013057 | G-A | HET | ||
| SBS | Chr4 | 1433380 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03340 |
| SBS | Chr4 | 30018834 | A-T | HET | ||
| SBS | Chr4 | 30018836 | C-T | HET | ||
| SBS | Chr4 | 34085242 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g57210 |
| SBS | Chr4 | 35117212 | G-A | HET | ||
| SBS | Chr4 | 4225103 | C-T | HET | ||
| SBS | Chr4 | 6545209 | G-A | Homo | ||
| SBS | Chr4 | 8497016 | G-A | HET | ||
| SBS | Chr4 | 8975938 | T-C | HET | ||
| SBS | Chr5 | 10454410 | C-T | Homo | ||
| SBS | Chr5 | 4655583 | G-A | HET | ||
| SBS | Chr6 | 2926067 | A-C | Homo | ||
| SBS | Chr7 | 13367718 | G-A | HET | ||
| SBS | Chr7 | 17505606 | T-A | HET | ||
| SBS | Chr7 | 19063756 | C-T | HET | ||
| SBS | Chr7 | 25483971 | A-G | HET | ||
| SBS | Chr8 | 18976841 | T-A | HET | ||
| SBS | Chr8 | 28044823 | C-T | HET | ||
| SBS | Chr8 | 3325507 | T-C | Homo | ||
| SBS | Chr8 | 6970840 | T-C | Homo | ||
| SBS | Chr8 | 7885733 | T-A | Homo | ||
| SBS | Chr8 | 7885734 | T-A | Homo | ||
| SBS | Chr9 | 4787324 | C-T | HET | ||
| SBS | Chr9 | 5536509 | A-T | Homo | ||
| SBS | Chr9 | 5682872 | C-T | HET | ||
| SBS | Chr9 | 7060859 | A-T | Homo |
Deletions: 16
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr7 | 2405365 | 2405374 | 9 | |
| Deletion | Chr7 | 2970162 | 2970168 | 6 | |
| Deletion | Chr5 | 2998650 | 2998652 | 2 | LOC_Os05g05990 |
| Deletion | Chr3 | 4915596 | 4915597 | 1 | |
| Deletion | Chr2 | 4963330 | 4963331 | 1 | |
| Deletion | Chr4 | 5291001 | 5326000 | 34999 | 4 |
| Deletion | Chr4 | 5328001 | 5365000 | 36999 | 6 |
| Deletion | Chr10 | 6497194 | 6497198 | 4 | LOC_Os10g11750 |
| Deletion | Chr9 | 6948274 | 6948277 | 3 | |
| Deletion | Chr9 | 9221226 | 9221227 | 1 | LOC_Os09g15190 |
| Deletion | Chr9 | 9302755 | 9302758 | 3 | |
| Deletion | Chr1 | 12893960 | 12893964 | 4 | |
| Deletion | Chr5 | 18845086 | 18845087 | 1 | LOC_Os05g32320 |
| Deletion | Chr2 | 23364222 | 23364223 | 1 | |
| Deletion | Chr8 | 27088482 | 27088497 | 15 | LOC_Os08g42840 |
| Deletion | Chr2 | 32226476 | 32226485 | 9 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 17347036 | 17347037 | 2 |
Inversions: 57
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 1471364 | 1471780 | LOC_Os08g03180 |
| Inversion | Chr8 | 1824024 | 1824178 | |
| Inversion | Chr4 | 1903953 | 1904251 | |
| Inversion | Chr10 | 2054791 | 2055170 | LOC_Os10g04342 |
| Inversion | Chr6 | 2370738 | 2370856 | LOC_Os06g05250 |
| Inversion | Chr6 | 2838298 | 2838417 | LOC_Os06g06130 |
| Inversion | Chr12 | 2932986 | 2933314 | LOC_Os12g06180 |
| Inversion | Chr7 | 3488748 | 3489003 | LOC_Os07g07070 |
| Inversion | Chr1 | 4022817 | 4023138 | |
| Inversion | Chr4 | 5292893 | 5659872 | 2 |
| Inversion | Chr7 | 6787816 | 6788022 | |
| Inversion | Chr3 | 6855353 | 6855482 | LOC_Os03g12790 |
| Inversion | Chr1 | 7106587 | 7106803 | LOC_Os01g12840 |
| Inversion | Chr6 | 8891126 | 8932760 | LOC_Os06g15700 |
| Inversion | Chr6 | 8891129 | 8932775 | LOC_Os06g15700 |
| Inversion | Chr12 | 10229473 | 10229897 | |
| Inversion | Chr10 | 10765936 | 10766443 | |
| Inversion | Chr6 | 12753301 | 12753565 | |
| Inversion | Chr5 | 12872723 | 12872840 | |
| Inversion | Chr10 | 13238656 | 13309155 | |
| Inversion | Chr10 | 13240217 | 13309154 | |
| Inversion | Chr3 | 14742920 | 14743114 | |
| Inversion | Chr1 | 15344159 | 15344578 | 2 |
| Inversion | Chr12 | 15454599 | 15454750 | |
| Inversion | Chr4 | 16588337 | 16588549 | |
| Inversion | Chr12 | 18831911 | 18832137 | LOC_Os12g31290 |
| Inversion | Chr1 | 19010187 | 19010473 | LOC_Os01g34480 |
| Inversion | Chr9 | 20442409 | 20538626 | 2 |
| Inversion | Chr9 | 20442425 | 20538641 | 2 |
| Inversion | Chr7 | 20639677 | 20640019 | |
| Inversion | Chr10 | 21884479 | 21884879 | LOC_Os10g40780 |
| Inversion | Chr11 | 22330150 | 22330379 | LOC_Os11g37759 |
| Inversion | Chr7 | 22809896 | 22810283 | |
| Inversion | Chr3 | 23318094 | 23318280 | LOC_Os03g41960 |
| Inversion | Chr4 | 24146345 | 24146486 | LOC_Os04g40680 |
| Inversion | Chr5 | 24254100 | 24254261 | |
| Inversion | Chr4 | 24567123 | 24567495 | LOC_Os04g41460 |
| Inversion | Chr11 | 25301395 | 25301680 | |
| Inversion | Chr5 | 25828223 | 25828622 | |
| Inversion | Chr4 | 26916154 | 27166624 | LOC_Os04g45480 |
| Inversion | Chr4 | 27717528 | 28217191 | 2 |
| Inversion | Chr4 | 27717550 | 27732635 | 2 |
| Inversion | Chr4 | 27718777 | 28346414 | |
| Inversion | Chr4 | 27719179 | 28346419 | |
| Inversion | Chr4 | 27732403 | 28217205 | 2 |
| Inversion | Chr4 | 28006309 | 28217211 | 2 |
| Inversion | Chr6 | 28413214 | 28413395 | |
| Inversion | Chr11 | 28690625 | 28690813 | |
| Inversion | Chr3 | 29580326 | 29580441 | |
| Inversion | Chr1 | 29843158 | 29843398 | LOC_Os01g51920 |
| Inversion | Chr1 | 31424212 | 31424719 | LOC_Os01g54620 |
| Inversion | Chr3 | 32622834 | 32623150 | |
| Inversion | Chr2 | 34045518 | 34045634 | LOC_Os02g55590 |
| Inversion | Chr2 | 35478405 | 35478548 | |
| Inversion | Chr3 | 36264928 | 36265213 | |
| Inversion | Chr1 | 37625886 | 37626296 | LOC_Os01g64820 |
| Inversion | Chr1 | 40443362 | 40443583 | LOC_Os01g69940 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr8 | 2793459 | Chr7 | 3174398 | |
| Translocation | Chr10 | 6480246 | Chr1 | 17210222 | |
| Translocation | Chr10 | 6480387 | Chr1 | 17210219 | LOC_Os10g11720 |
| Translocation | Chr7 | 8440488 | Chr4 | 28001169 | |
| Translocation | Chr4 | 17146872 | Chr1 | 17210222 | |
| Translocation | Chr11 | 18476138 | Chr10 | 6480581 | 2 |
| Translocation | Chr11 | 18476196 | Chr10 | 6480398 | 2 |
| Translocation | Chr11 | 28436075 | Chr1 | 27110217 | LOC_Os11g47290 |
