Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2137-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2137-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 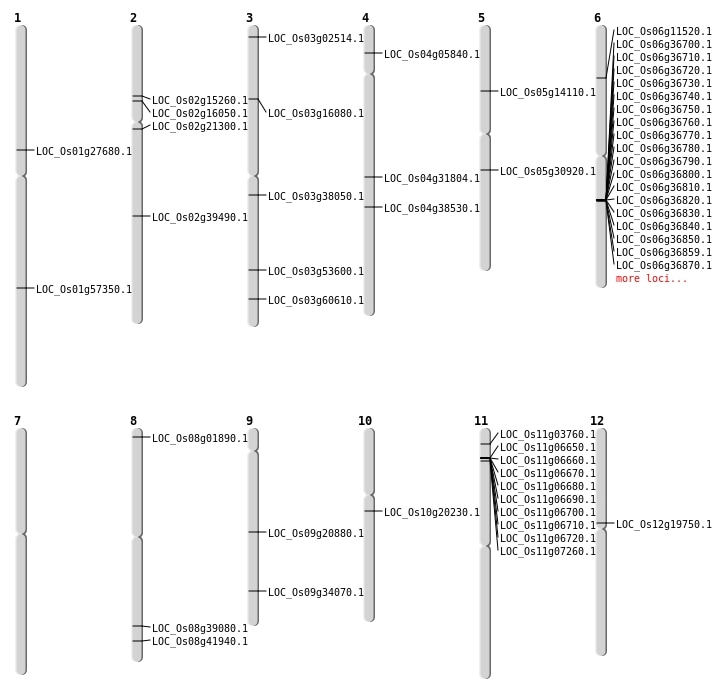
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14032035 | G-A | HET | ||
| SBS | Chr1 | 6355912 | A-T | HET | ||
| SBS | Chr10 | 15776797 | G-A | HET | ||
| SBS | Chr10 | 6633157 | C-T | HET | ||
| SBS | Chr11 | 16713043 | G-A | HET | ||
| SBS | Chr11 | 25447022 | A-G | Homo | ||
| SBS | Chr11 | 2631017 | C-T | HET | ||
| SBS | Chr12 | 11501707 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19750 |
| SBS | Chr12 | 12489681 | T-C | HET | ||
| SBS | Chr12 | 13848779 | C-A | HET | ||
| SBS | Chr12 | 17639760 | A-G | HET | ||
| SBS | Chr12 | 21775275 | C-T | HET | ||
| SBS | Chr12 | 4937880 | C-T | HET | ||
| SBS | Chr12 | 8248376 | G-A | HET | ||
| SBS | Chr2 | 12703416 | T-C | HET | ||
| SBS | Chr2 | 29653389 | G-A | HET | ||
| SBS | Chr2 | 9123788 | T-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g16050 |
| SBS | Chr3 | 1097091 | C-A | Homo | ||
| SBS | Chr4 | 11128363 | A-T | Homo | ||
| SBS | Chr4 | 20231960 | A-T | HET | ||
| SBS | Chr4 | 21519124 | C-T | Homo | ||
| SBS | Chr4 | 4911845 | C-T | Homo | ||
| SBS | Chr5 | 13026739 | T-C | Homo | ||
| SBS | Chr5 | 19885602 | C-A | Homo | ||
| SBS | Chr5 | 7884401 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os05g14110 |
| SBS | Chr6 | 7213385 | T-A | HET | ||
| SBS | Chr7 | 10624066 | G-A | Homo | ||
| SBS | Chr9 | 12566798 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os09g20880 |
| SBS | Chr9 | 16784844 | A-G | HET | ||
| SBS | Chr9 | 645194 | C-T | HET | ||
| SBS | Chr9 | 9296246 | G-T | Homo |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr11 | 3230001 | 3279000 | 48999 | 8 |
| Deletion | Chr4 | 8556027 | 8556028 | 1 | |
| Deletion | Chr8 | 9519548 | 9519549 | 1 | |
| Deletion | Chr2 | 9671204 | 9671205 | 1 | |
| Deletion | Chr11 | 12267300 | 12267407 | 107 | |
| Deletion | Chr2 | 12647254 | 12647255 | 1 | LOC_Os02g21300 |
| Deletion | Chr6 | 21602001 | 21767000 | 164999 | 26 |
| Deletion | Chr11 | 21678197 | 21678198 | 1 | |
| Deletion | Chr6 | 21797001 | 21847000 | 49999 | 4 |
| Deletion | Chr4 | 30511059 | 30511065 | 6 | |
| Deletion | Chr3 | 31015032 | 31015044 | 12 | |
| Deletion | Chr3 | 35921198 | 35921202 | 4 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 15059864 | 15059865 | 2 | |
| Insertion | Chr4 | 17081858 | 17081858 | 1 | |
| Insertion | Chr7 | 17560179 | 17560180 | 2 |
Inversions: 43
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr10 | 280308 | 280661 | |
| Inversion | Chr4 | 361887 | 362053 | |
| Inversion | Chr8 | 546955 | 547192 | LOC_Os08g01890 |
| Inversion | Chr12 | 792717 | 793056 | |
| Inversion | Chr3 | 928168 | 928504 | LOC_Os03g02514 |
| Inversion | Chr4 | 2033319 | 2033586 | |
| Inversion | Chr4 | 3002803 | 3003252 | LOC_Os04g05840 |
| Inversion | Chr5 | 3218382 | 3218764 | |
| Inversion | Chr3 | 3487631 | 3487750 | |
| Inversion | Chr11 | 3657839 | 3658394 | LOC_Os11g07260 |
| Inversion | Chr10 | 4561471 | 4561853 | |
| Inversion | Chr3 | 5991290 | 5991784 | |
| Inversion | Chr6 | 6104445 | 6104710 | LOC_Os06g11520 |
| Inversion | Chr7 | 6578338 | 6578812 | |
| Inversion | Chr9 | 6846496 | 6846731 | |
| Inversion | Chr2 | 8526682 | 8527002 | LOC_Os02g15260 |
| Inversion | Chr3 | 8866260 | 8866393 | LOC_Os03g16080 |
| Inversion | Chr8 | 8995824 | 8995944 | |
| Inversion | Chr9 | 9287898 | 9288189 | |
| Inversion | Chr9 | 9570254 | 9570538 | |
| Inversion | Chr10 | 10163544 | 10163769 | LOC_Os10g20230 |
| Inversion | Chr11 | 10565880 | 10566313 | |
| Inversion | Chr1 | 11860820 | 11861081 | |
| Inversion | Chr1 | 15439162 | 15439499 | LOC_Os01g27680 |
| Inversion | Chr12 | 17611484 | 17611833 | |
| Inversion | Chr5 | 17951955 | 17952487 | LOC_Os05g30920 |
| Inversion | Chr4 | 19056057 | 19056211 | LOC_Os04g31804 |
| Inversion | Chr9 | 20106752 | 20107066 | 2 |
| Inversion | Chr8 | 20395116 | 20395363 | |
| Inversion | Chr3 | 21122321 | 21122601 | LOC_Os03g38050 |
| Inversion | Chr6 | 23150409 | 23150681 | |
| Inversion | Chr2 | 23829850 | 23830115 | LOC_Os02g39490 |
| Inversion | Chr8 | 24689442 | 24689772 | LOC_Os08g39080 |
| Inversion | Chr8 | 26503078 | 26503414 | LOC_Os08g41940 |
| Inversion | Chr2 | 29269273 | 29269637 | |
| Inversion | Chr3 | 29608664 | 29609243 | |
| Inversion | Chr4 | 30003012 | 30003210 | |
| Inversion | Chr2 | 30565213 | 30565314 | |
| Inversion | Chr3 | 30737559 | 30737932 | LOC_Os03g53600 |
| Inversion | Chr3 | 32992033 | 32992320 | |
| Inversion | Chr1 | 33151271 | 33151523 | LOC_Os01g57350 |
| Inversion | Chr3 | 34442594 | 34442721 | LOC_Os03g60610 |
| Inversion | Chr1 | 42313706 | 42313857 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1475106 | Chr4 | 22897197 | 2 |
| Translocation | Chr12 | 7141995 | Chr11 | 13985386 | |
| Translocation | Chr12 | 7701218 | Chr1 | 8125310 | |
| Translocation | Chr12 | 7701746 | Chr1 | 8125275 |
