Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2150-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2150-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 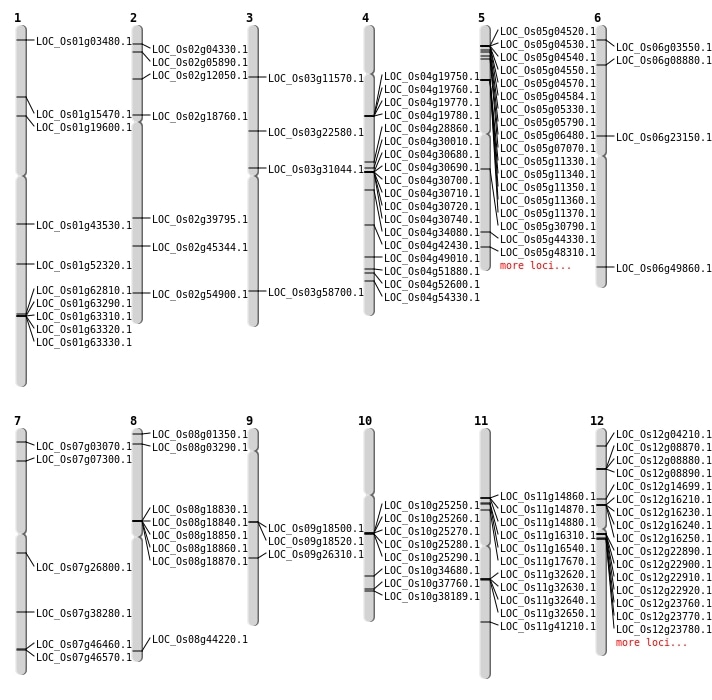
|
| Variant Information |
|---|
Single base substitutions: 27
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 42411713 | C-T | Homo | ||
| SBS | Chr10 | 15995517 | A-G | HET | ||
| SBS | Chr10 | 19692310 | C-T | HET | ||
| SBS | Chr10 | 22765201 | C-T | Homo | ||
| SBS | Chr10 | 4485508 | C-T | Homo | ||
| SBS | Chr11 | 15945416 | G-T | HET | ||
| SBS | Chr11 | 8728614 | C-T | HET | ||
| SBS | Chr12 | 24471858 | T-C | HET | ||
| SBS | Chr2 | 10721084 | T-C | HET | ||
| SBS | Chr2 | 21835254 | T-G | HET | ||
| SBS | Chr2 | 22011914 | G-A | HET | ||
| SBS | Chr2 | 2492540 | G-A | HET | ||
| SBS | Chr2 | 32282191 | G-A | HET | ||
| SBS | Chr2 | 33245815 | C-T | HET | ||
| SBS | Chr5 | 15488729 | C-G | HET | ||
| SBS | Chr5 | 1927427 | A-T | HET | ||
| SBS | Chr5 | 23633004 | T-C | HET | ||
| SBS | Chr5 | 25148910 | A-G | Homo | ||
| SBS | Chr6 | 12675771 | C-T | HET | ||
| SBS | Chr6 | 14632798 | G-A | HET | ||
| SBS | Chr6 | 24372117 | A-G | Homo | ||
| SBS | Chr6 | 30184602 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os06g49860 |
| SBS | Chr6 | 8787552 | C-T | HET | ||
| SBS | Chr7 | 17023946 | A-G | HET | ||
| SBS | Chr7 | 17631719 | G-T | HET | ||
| SBS | Chr8 | 17265504 | C-A | HET | ||
| SBS | Chr8 | 25436771 | C-T | HET |
Deletions: 29
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr12 | 983508 | 983510 | 2 | |
| Deletion | Chr11 | 1435267 | 1435275 | 8 | |
| Deletion | Chr5 | 2098001 | 2162000 | 63999 | 6 |
| Deletion | Chr10 | 4005487 | 4005488 | 1 | |
| Deletion | Chr11 | 4539113 | 4539116 | 3 | |
| Deletion | Chr12 | 4583001 | 4619000 | 35999 | 3 |
| Deletion | Chr5 | 6405001 | 6425000 | 19999 | 5 |
| Deletion | Chr11 | 8363001 | 8376000 | 12999 | 3 |
| Deletion | Chr6 | 8794697 | 8794701 | 4 | |
| Deletion | Chr11 | 8979001 | 9000000 | 20999 | LOC_Os11g16310 |
| Deletion | Chr12 | 9274001 | 9291000 | 16999 | 5 |
| Deletion | Chr1 | 10186488 | 10186490 | 2 | |
| Deletion | Chr4 | 11034001 | 11051000 | 16999 | 4 |
| Deletion | Chr1 | 11106837 | 11106839 | 2 | LOC_Os01g19600 |
| Deletion | Chr8 | 11239001 | 11262000 | 22999 | 5 |
| Deletion | Chr9 | 11338001 | 11358000 | 19999 | 2 |
| Deletion | Chr12 | 12924001 | 12956000 | 31999 | 4 |
| Deletion | Chr10 | 13043001 | 13070000 | 26999 | 5 |
| Deletion | Chr12 | 13497001 | 13554000 | 56999 | 8 |
| Deletion | Chr7 | 16200086 | 16200090 | 4 | |
| Deletion | Chr4 | 18335001 | 18377000 | 41999 | 6 |
| Deletion | Chr11 | 19241001 | 19280000 | 38999 | 4 |
| Deletion | Chr11 | 19351422 | 19351423 | 1 | |
| Deletion | Chr12 | 19974001 | 19997000 | 22999 | 3 |
| Deletion | Chr12 | 24261001 | 24279000 | 17999 | 2 |
| Deletion | Chr5 | 29590939 | 29590942 | 3 | LOC_Os05g51610 |
| Deletion | Chr1 | 36691001 | 36709000 | 17999 | 4 |
| Deletion | Chr1 | 39037029 | 39037034 | 5 | |
| Deletion | Chr1 | 39090984 | 39090985 | 1 |
Insertions: 6
Inversions: 115
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr8 | 223142 | 223631 | LOC_Os08g01350 |
| Inversion | Chr9 | 648886 | 649269 | |
| Inversion | Chr2 | 793475 | 793614 | |
| Inversion | Chr3 | 1162311 | 1162632 | |
| Inversion | Chr7 | 1167186 | 1167310 | LOC_Os07g03070 |
| Inversion | Chr6 | 1370847 | 1371097 | LOC_Os06g03550 |
| Inversion | Chr1 | 1401846 | 1402112 | LOC_Os01g03480 |
| Inversion | Chr8 | 1526676 | 1527029 | LOC_Os08g03290 |
| Inversion | Chr6 | 1731365 | 1731608 | |
| Inversion | Chr12 | 1776884 | 1777148 | LOC_Os12g04210 |
| Inversion | Chr2 | 1911730 | 1912022 | LOC_Os02g04330 |
| Inversion | Chr10 | 2238997 | 2239206 | |
| Inversion | Chr7 | 2410196 | 2410567 | |
| Inversion | Chr5 | 2627795 | 2627974 | LOC_Os05g05330 |
| Inversion | Chr5 | 2887460 | 2887846 | LOC_Os05g05790 |
| Inversion | Chr2 | 2923582 | 2923693 | LOC_Os02g05890 |
| Inversion | Chr5 | 3335680 | 3336079 | LOC_Os05g06480 |
| Inversion | Chr3 | 3366087 | 3366471 | |
| Inversion | Chr1 | 3414099 | 3414216 | |
| Inversion | Chr7 | 3628260 | 3628445 | LOC_Os07g07300 |
| Inversion | Chr5 | 3719617 | 3719926 | LOC_Os05g07070 |
| Inversion | Chr2 | 3976297 | 3976428 | |
| Inversion | Chr6 | 4462306 | 4462639 | LOC_Os06g08880 |
| Inversion | Chr1 | 5208319 | 5208602 | |
| Inversion | Chr4 | 5426072 | 5426218 | |
| Inversion | Chr11 | 5440369 | 5440538 | |
| Inversion | Chr3 | 6003558 | 6003879 | LOC_Os03g11570 |
| Inversion | Chr2 | 6259583 | 6259999 | LOC_Os02g12050 |
| Inversion | Chr6 | 6286322 | 6286544 | |
| Inversion | Chr2 | 6520006 | 6520364 | |
| Inversion | Chr5 | 6891005 | 6891166 | |
| Inversion | Chr6 | 6986838 | 6987075 | |
| Inversion | Chr4 | 8114169 | 8114524 | |
| Inversion | Chr12 | 8411119 | 8411520 | LOC_Os12g14699 |
| Inversion | Chr11 | 8596805 | 8596979 | |
| Inversion | Chr12 | 8660824 | 8661018 | |
| Inversion | Chr1 | 8685436 | 8685732 | LOC_Os01g15470 |
| Inversion | Chr11 | 9168263 | 9168464 | LOC_Os11g16540 |
| Inversion | Chr11 | 9862810 | 9862983 | LOC_Os11g17670 |
| Inversion | Chr4 | 10942650 | 10943105 | |
| Inversion | Chr3 | 12995208 | 12995357 | LOC_Os03g22580 |
| Inversion | Chr3 | 12998447 | 12998787 | LOC_Os03g22580 |
| Inversion | Chr6 | 13510091 | 13510424 | 2 |
| Inversion | Chr4 | 13551203 | 13551443 | |
| Inversion | Chr3 | 13639564 | 13639822 | |
| Inversion | Chr10 | 13984873 | 13984978 | |
| Inversion | Chr5 | 14244642 | 14244993 | |
| Inversion | Chr12 | 14492634 | 14493074 | |
| Inversion | Chr10 | 14685511 | 14685772 | |
| Inversion | Chr7 | 15041515 | 15041645 | |
| Inversion | Chr12 | 15288434 | 15288633 | |
| Inversion | Chr7 | 15304292 | 15489371 | 2 |
| Inversion | Chr7 | 15304298 | 15489375 | 2 |
| Inversion | Chr7 | 15882498 | 15882668 | |
| Inversion | Chr9 | 15896487 | 15896690 | LOC_Os09g26310 |
| Inversion | Chr10 | 16572852 | 16573029 | |
| Inversion | Chr4 | 16607771 | 16608017 | |
| Inversion | Chr4 | 17103456 | 17103657 | LOC_Os04g28860 |
| Inversion | Chr3 | 17686329 | 17686586 | LOC_Os03g31044 |
| Inversion | Chr5 | 17860227 | 17860499 | LOC_Os05g30790 |
| Inversion | Chr4 | 17906696 | 18052674 | LOC_Os04g30010 |
| Inversion | Chr10 | 18495593 | 18495910 | LOC_Os10g34680 |
| Inversion | Chr9 | 18992070 | 18992379 | |
| Inversion | Chr5 | 19091346 | 19091723 | |
| Inversion | Chr2 | 20084737 | 20084959 | |
| Inversion | Chr10 | 20208830 | 20209177 | LOC_Os10g37760 |
| Inversion | Chr7 | 20363539 | 20363657 | |
| Inversion | Chr10 | 20449209 | 20449370 | LOC_Os10g38189 |
| Inversion | Chr4 | 20641335 | 20641560 | LOC_Os04g34080 |
| Inversion | Chr7 | 20727435 | 20727741 | |
| Inversion | Chr11 | 20959964 | 20960153 | |
| Inversion | Chr12 | 21751161 | 21751373 | |
| Inversion | Chr10 | 22092647 | 22092832 | |
| Inversion | Chr9 | 22212018 | 22212152 | |
| Inversion | Chr6 | 22278598 | 22278713 | |
| Inversion | Chr7 | 22981528 | 22981761 | LOC_Os07g38280 |
| Inversion | Chr4 | 23004050 | 23004265 | |
| Inversion | Chr7 | 23993936 | 23994059 | |
| Inversion | Chr2 | 24048714 | 24049016 | LOC_Os02g39795 |
| Inversion | Chr7 | 24705350 | 24705662 | |
| Inversion | Chr11 | 24716157 | 24716434 | LOC_Os11g41210 |
| Inversion | Chr1 | 24932536 | 24932906 | 2 |
| Inversion | Chr4 | 25113464 | 25113890 | LOC_Os04g42430 |
| Inversion | Chr4 | 25496472 | 25496604 | |
| Inversion | Chr8 | 25630445 | 25630831 | |
| Inversion | Chr2 | 25693408 | 25693821 | |
| Inversion | Chr5 | 25801304 | 25801782 | LOC_Os05g44330 |
| Inversion | Chr8 | 26370921 | 26371286 | |
| Inversion | Chr2 | 26474926 | 26475206 | |
| Inversion | Chr8 | 26729569 | 26729945 | |
| Inversion | Chr11 | 26762804 | 26763122 | |
| Inversion | Chr7 | 27025279 | 27025444 | |
| Inversion | Chr5 | 27102743 | 27102885 | |
| Inversion | Chr3 | 27147847 | 27148224 | |
| Inversion | Chr2 | 27566485 | 27566709 | LOC_Os02g45344 |
| Inversion | Chr5 | 27697013 | 27697386 | LOC_Os05g48310 |
| Inversion | Chr7 | 27737419 | 27737610 | LOC_Os07g46460 |
| Inversion | Chr7 | 27813672 | 27814161 | 2 |
| Inversion | Chr1 | 28603686 | 28603938 | |
| Inversion | Chr6 | 29186351 | 29186512 | |
| Inversion | Chr4 | 29220566 | 29221117 | LOC_Os04g49010 |
| Inversion | Chr5 | 29498224 | 29498528 | LOC_Os05g51450 |
| Inversion | Chr6 | 30032419 | 30032939 | |
| Inversion | Chr1 | 30069041 | 30069499 | LOC_Os01g52320 |
| Inversion | Chr4 | 30767116 | 30767558 | LOC_Os04g51880 |
| Inversion | Chr1 | 30954914 | 30955109 | |
| Inversion | Chr4 | 31278094 | 31278363 | LOC_Os04g52600 |
| Inversion | Chr4 | 32339898 | 32340052 | LOC_Os04g54330 |
| Inversion | Chr2 | 32774694 | 32775109 | |
| Inversion | Chr1 | 33337126 | 33337610 | |
| Inversion | Chr3 | 33431037 | 33431585 | LOC_Os03g58700 |
| Inversion | Chr2 | 33627262 | 33627545 | LOC_Os02g54900 |
| Inversion | Chr1 | 36380750 | 36380859 | LOC_Os01g62810 |
| Inversion | Chr1 | 42176095 | 42176512 | |
| Inversion | Chr1 | 42943847 | 42944125 |
Translocations: 6
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 8941257 | Chr1 | 30237136 | |
| Translocation | Chr9 | 9611211 | Chr1 | 15261178 | |
| Translocation | Chr10 | 13647195 | Chr2 | 10937241 | 2 |
| Translocation | Chr12 | 19884557 | Chr8 | 27836248 | 2 |
| Translocation | Chr11 | 19917137 | Chr2 | 19309431 | |
| Translocation | Chr11 | 26712318 | Chr1 | 15324159 |
