Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2161-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2161-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 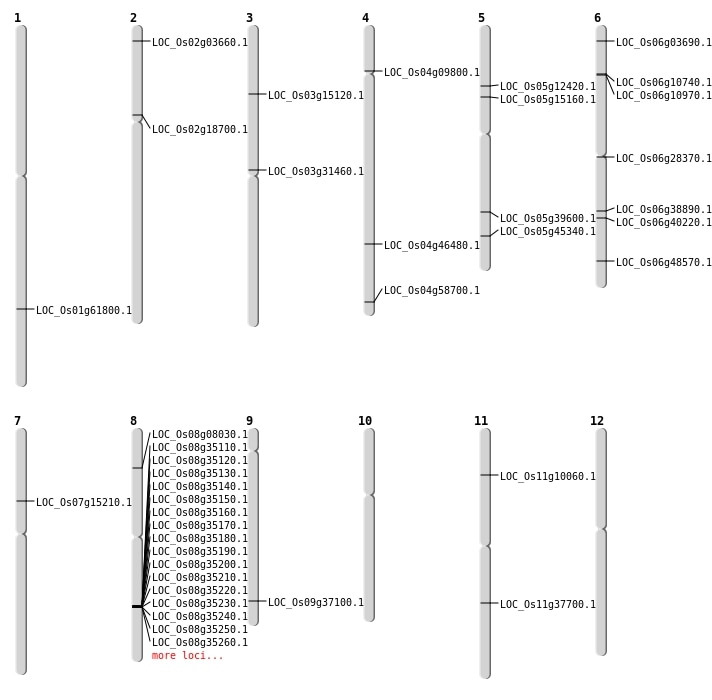
|
| Variant Information |
|---|
Single base substitutions: 31
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 11744249 | C-T | HET | ||
| SBS | Chr1 | 2482963 | A-C | HET | ||
| SBS | Chr1 | 36408575 | C-T | HET | ||
| SBS | Chr10 | 11060663 | C-T | HET | ||
| SBS | Chr10 | 6406642 | G-C | Homo | ||
| SBS | Chr11 | 1195730 | G-A | HET | ||
| SBS | Chr11 | 1549188 | A-G | HET | ||
| SBS | Chr11 | 18755744 | C-A | HET | ||
| SBS | Chr11 | 8679484 | A-G | Homo | ||
| SBS | Chr12 | 1600648 | C-T | Homo | ||
| SBS | Chr12 | 25989445 | A-T | HET | ||
| SBS | Chr2 | 10913190 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g18700 |
| SBS | Chr2 | 1525918 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os02g03660 |
| SBS | Chr3 | 13454491 | G-C | HET | ||
| SBS | Chr3 | 1625011 | G-T | Homo | ||
| SBS | Chr3 | 30059259 | T-C | HET | ||
| SBS | Chr3 | 8225247 | T-C | HET | ||
| SBS | Chr4 | 19022365 | T-A | HET | ||
| SBS | Chr4 | 33727319 | T-C | HET | ||
| SBS | Chr5 | 18558427 | G-A | HET | ||
| SBS | Chr5 | 21486229 | G-T | HET | ||
| SBS | Chr5 | 9003952 | C-T | HET | ||
| SBS | Chr6 | 14652885 | G-A | Homo | ||
| SBS | Chr6 | 23074349 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os06g38890 |
| SBS | Chr6 | 5733028 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g10970 |
| SBS | Chr7 | 14861586 | C-T | Homo | ||
| SBS | Chr7 | 24689586 | C-T | HET | ||
| SBS | Chr7 | 4810883 | G-A | Homo | ||
| SBS | Chr9 | 12674683 | G-A | HET | ||
| SBS | Chr9 | 12986443 | T-A | HET | ||
| SBS | Chr9 | 9766932 | C-T | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr6 | 3672689 | 3672692 | 3 | |
| Deletion | Chr6 | 5610574 | 5610575 | 1 | LOC_Os06g10740 |
| Deletion | Chr1 | 12400394 | 12400398 | 4 | |
| Deletion | Chr12 | 13289451 | 13289452 | 1 | |
| Deletion | Chr8 | 22128001 | 22167000 | 38999 | 7 |
| Deletion | Chr8 | 22171001 | 22322000 | 150999 | 24 |
| Deletion | Chr12 | 24587178 | 24587193 | 15 | |
| Deletion | Chr1 | 26939874 | 26939876 | 2 | |
| Deletion | Chr1 | 34848700 | 34848708 | 8 | |
| Deletion | Chr3 | 35839871 | 35839872 | 1 | |
| Deletion | Chr1 | 36907333 | 36907335 | 2 |
Insertions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 12434813 | 12434814 | 2 | |
| Insertion | Chr11 | 21581488 | 21581488 | 1 | |
| Insertion | Chr2 | 3574739 | 3574739 | 1 | |
| Insertion | Chr2 | 3601055 | 3601056 | 2 | |
| Insertion | Chr3 | 16565733 | 16565734 | 2 | |
| Insertion | Chr3 | 9570690 | 9570727 | 38 | |
| Insertion | Chr7 | 8766718 | 8766718 | 1 | LOC_Os07g15210 |
| Insertion | Chr8 | 7269366 | 7269366 | 1 | |
| Insertion | Chr8 | 8092507 | 8092507 | 1 | |
| Insertion | Chr9 | 13258549 | 13258549 | 1 | |
| Insertion | Chr9 | 2169452 | 2169455 | 4 |
Inversions: 28
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 198590 | 199012 | |
| Inversion | Chr8 | 544118 | 544603 | |
| Inversion | Chr6 | 1456288 | 1456556 | LOC_Os06g03690 |
| Inversion | Chr6 | 3970185 | 3970462 | |
| Inversion | Chr8 | 4553319 | 4553502 | LOC_Os08g08030 |
| Inversion | Chr10 | 4876888 | 4877175 | |
| Inversion | Chr4 | 5254916 | 5255105 | LOC_Os04g09800 |
| Inversion | Chr11 | 5400994 | 5401296 | LOC_Os11g10060 |
| Inversion | Chr5 | 6821704 | 6822002 | |
| Inversion | Chr5 | 7140642 | 7140844 | LOC_Os05g12420 |
| Inversion | Chr5 | 8607722 | 8607838 | LOC_Os05g15160 |
| Inversion | Chr5 | 9181262 | 9181367 | |
| Inversion | Chr4 | 13814183 | 13814400 | |
| Inversion | Chr6 | 16133973 | 16134093 | LOC_Os06g28370 |
| Inversion | Chr12 | 17794007 | 17794399 | |
| Inversion | Chr3 | 17931441 | 17931563 | LOC_Os03g31460 |
| Inversion | Chr10 | 18819872 | 18820018 | |
| Inversion | Chr9 | 21397362 | 21397479 | LOC_Os09g37100 |
| Inversion | Chr10 | 21525741 | 21525930 | |
| Inversion | Chr11 | 22275303 | 22275465 | LOC_Os11g37700 |
| Inversion | Chr5 | 23252120 | 23252276 | LOC_Os05g39600 |
| Inversion | Chr6 | 23956102 | 23956297 | 2 |
| Inversion | Chr6 | 23956674 | 23957138 | LOC_Os06g40220 |
| Inversion | Chr5 | 26303689 | 26304012 | LOC_Os05g45340 |
| Inversion | Chr4 | 27571956 | 27572173 | LOC_Os04g46480 |
| Inversion | Chr6 | 29384429 | 29384730 | LOC_Os06g48570 |
| Inversion | Chr1 | 29877758 | 29877919 | |
| Inversion | Chr1 | 35752743 | 35752916 | LOC_Os01g61800 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr3 | 8255468 | Chr2 | 14361985 | LOC_Os03g15120 |
| Translocation | Chr2 | 14983934 | Chr1 | 20917486 | |
| Translocation | Chr9 | 18095053 | Chr4 | 34904195 | 2 |
| Translocation | Chr8 | 27530336 | Chr3 | 31848814 | LOC_Os08g43530 |
