Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2177-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2177-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 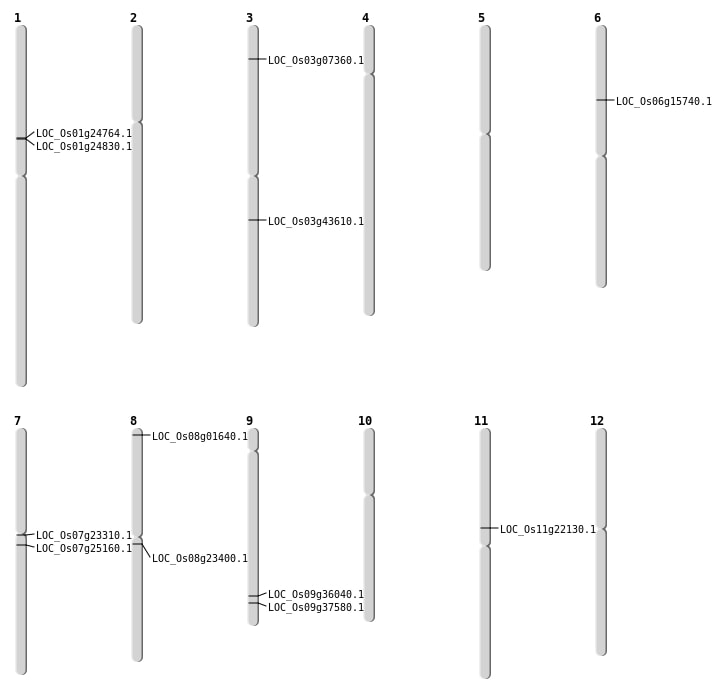
|
| Variant Information |
|---|
Single base substitutions: 41
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13985196 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os01g24830 |
| SBS | Chr1 | 23405264 | A-G | Homo | ||
| SBS | Chr1 | 25173579 | A-G | HET | ||
| SBS | Chr1 | 37312258 | C-A | HET | ||
| SBS | Chr1 | 6422101 | T-C | Homo | ||
| SBS | Chr1 | 8474087 | A-G | HET | ||
| SBS | Chr10 | 21101407 | C-T | Homo | ||
| SBS | Chr11 | 15364989 | G-A | Homo | ||
| SBS | Chr11 | 18836817 | A-G | HET | ||
| SBS | Chr12 | 12225040 | C-T | HET | ||
| SBS | Chr2 | 11747171 | T-A | Homo | ||
| SBS | Chr2 | 22682705 | G-A | HET | ||
| SBS | Chr2 | 24924695 | G-C | HET | ||
| SBS | Chr3 | 24374368 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g43610 |
| SBS | Chr3 | 25468794 | T-A | HET | ||
| SBS | Chr3 | 30879209 | T-G | HET | ||
| SBS | Chr3 | 35757661 | G-A | HET | ||
| SBS | Chr3 | 3740819 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g07360 |
| SBS | Chr3 | 6506541 | G-A | Homo | ||
| SBS | Chr3 | 973728 | A-T | Homo | ||
| SBS | Chr3 | 973729 | C-T | Homo | ||
| SBS | Chr4 | 10079951 | C-A | HET | ||
| SBS | Chr4 | 12704001 | C-A | HET | ||
| SBS | Chr4 | 13294847 | G-A | HET | ||
| SBS | Chr4 | 16435889 | T-A | Homo | ||
| SBS | Chr4 | 27839632 | T-G | HET | ||
| SBS | Chr4 | 28384018 | A-G | HET | ||
| SBS | Chr5 | 18584896 | C-T | HET | ||
| SBS | Chr6 | 10775255 | G-A | HET | ||
| SBS | Chr6 | 28962962 | G-A | HET | ||
| SBS | Chr6 | 28962967 | C-T | HET | ||
| SBS | Chr6 | 9122107 | C-T | HET | ||
| SBS | Chr7 | 13140680 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g23310 |
| SBS | Chr7 | 1543994 | G-A | HET | ||
| SBS | Chr7 | 26500129 | A-G | HET | ||
| SBS | Chr7 | 27672987 | C-G | HET | ||
| SBS | Chr7 | 8075024 | A-T | HET | ||
| SBS | Chr8 | 12664729 | G-C | HET | ||
| SBS | Chr8 | 26366926 | G-A | Homo | ||
| SBS | Chr8 | 940896 | G-T | HET | ||
| SBS | Chr9 | 15837953 | G-C | Homo |
Deletions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr8 | 374411 | 374424 | 13 | LOC_Os08g01640 |
| Deletion | Chr3 | 3714253 | 3714254 | 1 | |
| Deletion | Chr3 | 4742597 | 4742602 | 5 | |
| Deletion | Chr5 | 12019469 | 12019470 | 1 | |
| Deletion | Chr8 | 14144239 | 14144241 | 2 | LOC_Os08g23400 |
| Deletion | Chr4 | 15908757 | 15908765 | 8 | |
| Deletion | Chr1 | 16312140 | 16312151 | 11 | |
| Deletion | Chr5 | 21642613 | 21642624 | 11 | |
| Deletion | Chr9 | 21658213 | 21658215 | 2 | LOC_Os09g37580 |
| Deletion | Chr8 | 23621782 | 23621786 | 4 | |
| Deletion | Chr1 | 32690811 | 32690812 | 1 | |
| Deletion | Chr4 | 33433597 | 33433605 | 8 |
Insertions: 12
Inversions: 7
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 2066345 | 2368871 | |
| Inversion | Chr1 | 2066363 | 2368878 | |
| Inversion | Chr2 | 2249683 | 2249864 | |
| Inversion | Chr4 | 7933011 | 7933196 | |
| Inversion | Chr9 | 20752711 | 21138749 | LOC_Os09g36040 |
| Inversion | Chr9 | 20752726 | 21138755 | LOC_Os09g36040 |
| Inversion | Chr9 | 21138954 | 21139385 |
Translocations: 8
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr6 | 8926733 | Chr4 | 35190186 | LOC_Os06g15740 |
| Translocation | Chr6 | 8927009 | Chr4 | 35190192 | LOC_Os06g15740 |
| Translocation | Chr11 | 12699276 | Chr7 | 14859439 | LOC_Os11g22130 |
| Translocation | Chr11 | 12699374 | Chr3 | 23240930 | LOC_Os11g22130 |
| Translocation | Chr12 | 17941543 | Chr6 | 8926887 | 2 |
| Translocation | Chr12 | 19376830 | Chr1 | 13934750 | 2 |
| Translocation | Chr9 | 21853392 | Chr7 | 14380603 | 2 |
| Translocation | Chr9 | 21853399 | Chr7 | 14372271 |
