Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2181-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2181-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 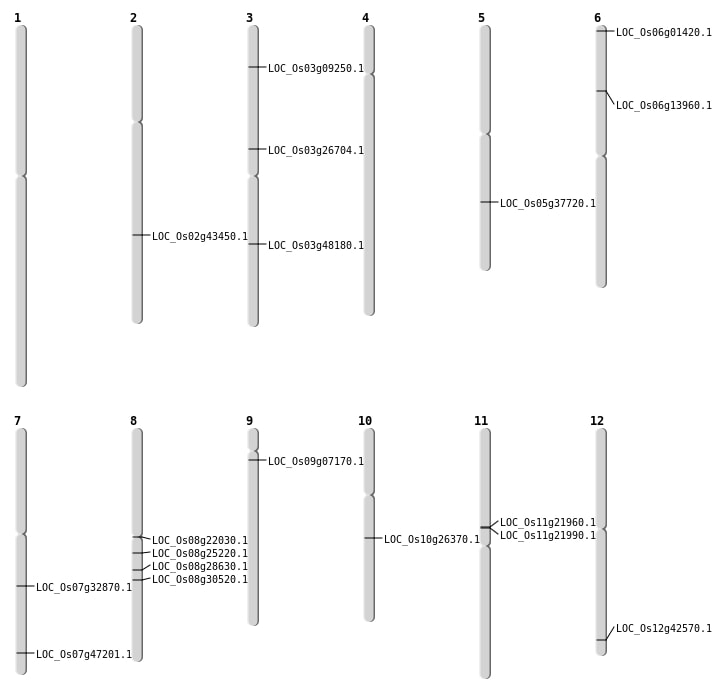
|
| Variant Information |
|---|
Single base substitutions: 45
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 29857391 | G-T | Homo | ||
| SBS | Chr1 | 8714329 | A-T | HET | ||
| SBS | Chr10 | 22845938 | C-A | HET | ||
| SBS | Chr10 | 4639384 | G-T | HET | ||
| SBS | Chr11 | 10455773 | C-G | HET | ||
| SBS | Chr11 | 28927758 | G-A | HET | ||
| SBS | Chr11 | 4759739 | A-C | HET | ||
| SBS | Chr12 | 19989171 | A-G | HET | ||
| SBS | Chr12 | 20323249 | T-A | Homo | ||
| SBS | Chr12 | 22201052 | C-T | HET | ||
| SBS | Chr12 | 26435119 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g42570 |
| SBS | Chr12 | 2891569 | T-A | HET | ||
| SBS | Chr2 | 12200367 | G-A | HET | ||
| SBS | Chr2 | 30069553 | A-C | Homo | ||
| SBS | Chr2 | 30088814 | C-T | Homo | ||
| SBS | Chr3 | 27414728 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g48180 |
| SBS | Chr3 | 27414729 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g48180 |
| SBS | Chr3 | 29419488 | C-A | HET | ||
| SBS | Chr3 | 32296860 | G-T | HET | ||
| SBS | Chr3 | 32908937 | G-A | HET | ||
| SBS | Chr3 | 4826646 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g09250 |
| SBS | Chr3 | 6221470 | T-A | HET | ||
| SBS | Chr4 | 10886749 | T-C | HET | ||
| SBS | Chr4 | 17235393 | A-T | HET | ||
| SBS | Chr4 | 24291015 | T-C | Homo | ||
| SBS | Chr4 | 25834734 | C-T | Homo | ||
| SBS | Chr4 | 7977785 | T-G | HET | ||
| SBS | Chr5 | 16890867 | T-A | Homo | ||
| SBS | Chr5 | 21335322 | A-G | HET | ||
| SBS | Chr5 | 21335333 | C-G | HET | ||
| SBS | Chr5 | 22076862 | T-C | Homo | NON_SYNONYMOUS_CODING | LOC_Os05g37720 |
| SBS | Chr5 | 23144770 | T-C | HET | ||
| SBS | Chr5 | 7150376 | G-A | Homo | ||
| SBS | Chr6 | 255820 | C-T | HET | STOP_GAINED | LOC_Os06g01420 |
| SBS | Chr6 | 4005220 | T-A | HET | ||
| SBS | Chr7 | 19654882 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g32870 |
| SBS | Chr7 | 27198245 | C-A | Homo | ||
| SBS | Chr7 | 7377206 | C-T | HET | ||
| SBS | Chr8 | 13306338 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os08g22030 |
| SBS | Chr8 | 18777246 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g30520 |
| SBS | Chr8 | 5933665 | G-T | HET | ||
| SBS | Chr9 | 1716936 | G-A | HET | ||
| SBS | Chr9 | 1971522 | C-G | HET | ||
| SBS | Chr9 | 3513546 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os09g07170 |
| SBS | Chr9 | 5168412 | G-A | HET |
Deletions: 11
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1969247 | 1969252 | 5 | |
| Deletion | Chr12 | 2582191 | 2582195 | 4 | |
| Deletion | Chr10 | 8469016 | 8469019 | 3 | |
| Deletion | Chr2 | 11471995 | 11471999 | 4 | |
| Deletion | Chr10 | 13687951 | 13687952 | 1 | LOC_Os10g26370 |
| Deletion | Chr1 | 14347401 | 14347402 | 1 | |
| Deletion | Chr12 | 22946465 | 22946467 | 2 | |
| Deletion | Chr1 | 23434435 | 23434439 | 4 | |
| Deletion | Chr2 | 26223411 | 26223416 | 5 | LOC_Os02g43450 |
| Deletion | Chr8 | 26701221 | 26701223 | 2 | |
| Deletion | Chr1 | 28356939 | 28356945 | 6 |
No Insertion
Inversions: 3
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr11 | 12599609 | 12633603 | 2 |
| Inversion | Chr6 | 25511163 | 25679790 | |
| Inversion | Chr6 | 25511164 | 25679792 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 11379209 | Chr6 | 7774338 | 2 |
| Translocation | Chr8 | 15226187 | Chr1 | 2638277 | |
| Translocation | Chr8 | 15325646 | Chr4 | 5328030 | LOC_Os08g25220 |
| Translocation | Chr8 | 17495802 | Chr3 | 15244343 | 2 |
| Translocation | Chr7 | 28216500 | Chr4 | 17793219 | LOC_Os07g47201 |
