Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2183-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2183-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 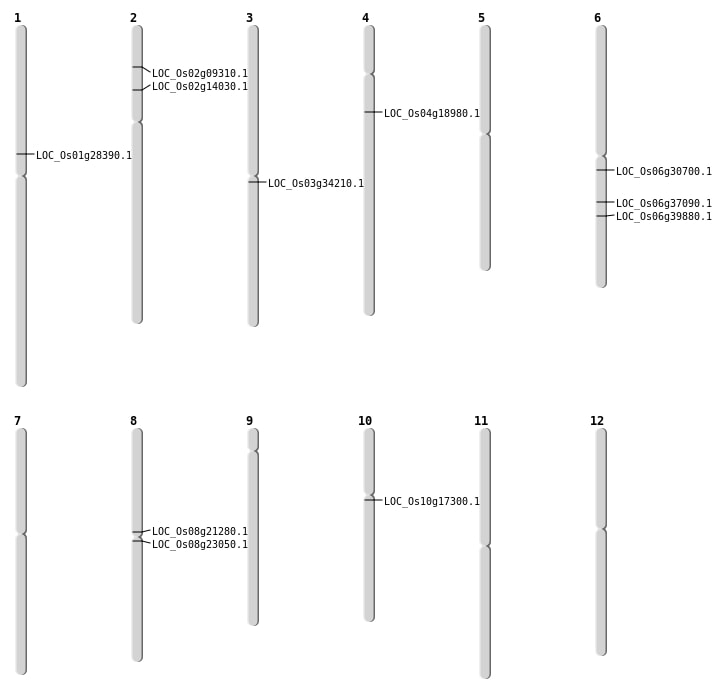
|
| Variant Information |
|---|
Single base substitutions: 37
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 13198556 | G-C | Homo | ||
| SBS | Chr1 | 15910192 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os01g28390 |
| SBS | Chr1 | 16533318 | A-C | HET | ||
| SBS | Chr1 | 25636344 | G-A | HET | ||
| SBS | Chr1 | 43243633 | C-G | HET | ||
| SBS | Chr1 | 8000123 | C-G | HET | ||
| SBS | Chr1 | 9003112 | A-C | HET | ||
| SBS | Chr10 | 17767966 | T-C | Homo | ||
| SBS | Chr10 | 6822471 | C-A | HET | ||
| SBS | Chr10 | 8705394 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g17300 |
| SBS | Chr11 | 1760742 | G-A | HET | ||
| SBS | Chr11 | 24946161 | C-T | Homo | ||
| SBS | Chr12 | 18196687 | T-C | HET | ||
| SBS | Chr12 | 27207326 | C-A | HET | ||
| SBS | Chr12 | 3726138 | G-A | HET | ||
| SBS | Chr2 | 12296959 | T-C | HET | ||
| SBS | Chr2 | 1288988 | A-T | HET | ||
| SBS | Chr2 | 2425897 | C-T | HET | ||
| SBS | Chr2 | 30277452 | G-T | Homo | ||
| SBS | Chr2 | 31871352 | G-A | Homo | ||
| SBS | Chr2 | 4790469 | T-A | HET | ||
| SBS | Chr2 | 73021 | T-C | Homo | ||
| SBS | Chr2 | 7658757 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g14030 |
| SBS | Chr3 | 19481881 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os03g34210 |
| SBS | Chr3 | 33970520 | G-A | HET | ||
| SBS | Chr3 | 34026811 | C-T | HET | ||
| SBS | Chr4 | 15479105 | G-A | HET | ||
| SBS | Chr5 | 20451664 | C-T | HET | ||
| SBS | Chr5 | 24896281 | C-A | HET | ||
| SBS | Chr5 | 7498733 | C-T | HET | ||
| SBS | Chr6 | 12032929 | A-T | HET | ||
| SBS | Chr6 | 17809084 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g30700 |
| SBS | Chr7 | 16727503 | G-A | HET | ||
| SBS | Chr8 | 10701677 | C-T | HET | ||
| SBS | Chr8 | 12701925 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21280 |
| SBS | Chr8 | 636722 | C-T | HET | ||
| SBS | Chr9 | 15760867 | G-A | HET |
Deletions: 33
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr4 | 433213 | 433215 | 2 | |
| Deletion | Chr5 | 1090098 | 1090099 | 1 | |
| Deletion | Chr3 | 2483553 | 2483554 | 1 | |
| Deletion | Chr5 | 4259239 | 4259240 | 1 | |
| Deletion | Chr12 | 4522099 | 4522113 | 14 | |
| Deletion | Chr2 | 4792348 | 4792360 | 12 | LOC_Os02g09310 |
| Deletion | Chr4 | 5966765 | 5966773 | 8 | |
| Deletion | Chr9 | 6930851 | 6930854 | 3 | |
| Deletion | Chr3 | 6939778 | 6939788 | 10 | |
| Deletion | Chr12 | 9749684 | 9749692 | 8 | |
| Deletion | Chr4 | 10547092 | 10547110 | 18 | LOC_Os04g18980 |
| Deletion | Chr2 | 11679612 | 11679615 | 3 | |
| Deletion | Chr11 | 13042658 | 13042667 | 9 | |
| Deletion | Chr5 | 13075588 | 13075593 | 5 | |
| Deletion | Chr2 | 13599669 | 13599679 | 10 | |
| Deletion | Chr8 | 13873256 | 13873270 | 14 | LOC_Os08g23050 |
| Deletion | Chr8 | 14415673 | 14415676 | 3 | |
| Deletion | Chr2 | 15507345 | 15507350 | 5 | |
| Deletion | Chr10 | 17456314 | 17456318 | 4 | |
| Deletion | Chr10 | 17821577 | 17821586 | 9 | |
| Deletion | Chr3 | 18888532 | 18888538 | 6 | |
| Deletion | Chr3 | 20198288 | 20198299 | 11 | |
| Deletion | Chr6 | 21907267 | 21907279 | 12 | LOC_Os06g37090 |
| Deletion | Chr1 | 23695705 | 23695716 | 11 | |
| Deletion | Chr6 | 23705724 | 23705734 | 10 | LOC_Os06g39880 |
| Deletion | Chr1 | 23823683 | 23823691 | 8 | |
| Deletion | Chr1 | 25636322 | 25636333 | 11 | |
| Deletion | Chr4 | 29213597 | 29213600 | 3 | |
| Deletion | Chr1 | 30114090 | 30114091 | 1 | |
| Deletion | Chr3 | 30149266 | 30149271 | 5 | |
| Deletion | Chr3 | 32633078 | 32633085 | 7 | |
| Deletion | Chr2 | 34214506 | 34214513 | 7 | |
| Deletion | Chr2 | 34688089 | 34688099 | 10 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr11 | 20304055 | 20304060 | 6 | |
| Insertion | Chr12 | 26316418 | 26316419 | 2 | |
| Insertion | Chr3 | 98887 | 98888 | 2 | |
| Insertion | Chr4 | 3726413 | 3726413 | 1 |
No Inversion
Translocations: 2
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr9 | 7362307 | Chr2 | 28410003 | |
| Translocation | Chr5 | 29808250 | Chr4 | 2440064 |
