Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2191-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2191-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 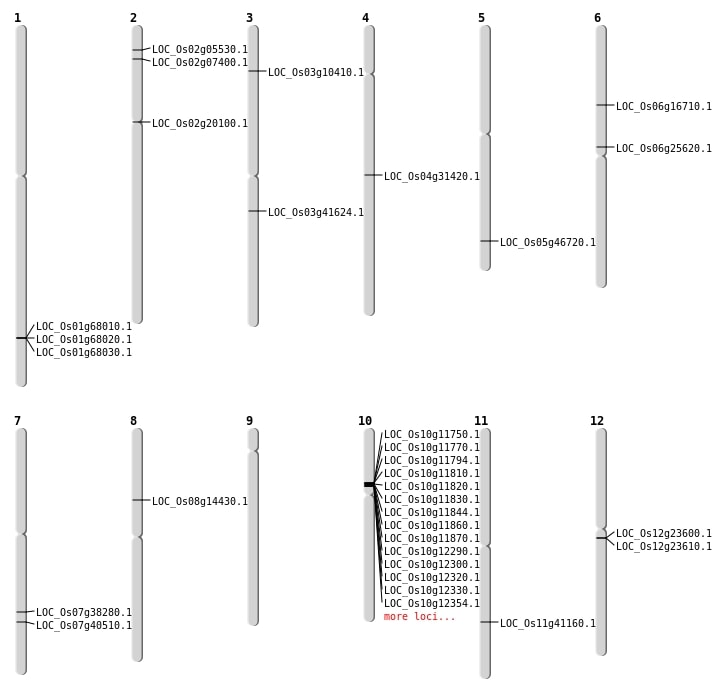
|
| Variant Information |
|---|
Single base substitutions: 46
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 22774857 | T-C | Homo | ||
| SBS | Chr1 | 28310334 | G-T | HET | ||
| SBS | Chr1 | 38100905 | T-C | HET | ||
| SBS | Chr1 | 38730251 | T-C | HET | ||
| SBS | Chr1 | 39380044 | C-T | HET | ||
| SBS | Chr1 | 6641879 | A-G | HET | ||
| SBS | Chr1 | 8730592 | A-T | HET | ||
| SBS | Chr10 | 11326779 | G-C | HET | ||
| SBS | Chr10 | 13572712 | C-T | HET | ||
| SBS | Chr10 | 8901157 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g17610 |
| SBS | Chr11 | 21779356 | A-T | Homo | ||
| SBS | Chr11 | 21779357 | A-T | Homo | ||
| SBS | Chr11 | 21779358 | A-T | Homo | ||
| SBS | Chr11 | 23532492 | C-G | HET | ||
| SBS | Chr11 | 9521397 | G-C | Homo | ||
| SBS | Chr12 | 6536098 | G-T | HET | ||
| SBS | Chr2 | 11837043 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g20100 |
| SBS | Chr2 | 17296219 | A-G | HET | ||
| SBS | Chr2 | 3822100 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os02g07400 |
| SBS | Chr3 | 11756185 | G-A | Homo | ||
| SBS | Chr3 | 25516387 | A-G | HET | ||
| SBS | Chr3 | 35144541 | T-C | HET | ||
| SBS | Chr3 | 5302499 | A-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os03g10410 |
| SBS | Chr4 | 12565977 | C-T | HET | ||
| SBS | Chr4 | 16772008 | G-A | HET | ||
| SBS | Chr4 | 17701642 | G-T | HET | ||
| SBS | Chr4 | 18497568 | C-T | HET | ||
| SBS | Chr4 | 18793511 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g31420 |
| SBS | Chr5 | 22318718 | C-T | Homo | ||
| SBS | Chr5 | 29751673 | G-A | HET | ||
| SBS | Chr6 | 12335538 | T-C | HET | ||
| SBS | Chr6 | 14982660 | C-T | HET | STOP_GAINED | LOC_Os06g25620 |
| SBS | Chr6 | 22226898 | A-G | HET | ||
| SBS | Chr6 | 25573534 | C-T | HET | ||
| SBS | Chr6 | 25744001 | A-T | HET | ||
| SBS | Chr7 | 17966795 | C-T | Homo | ||
| SBS | Chr7 | 22684518 | G-A | Homo | ||
| SBS | Chr7 | 24272386 | A-C | HET | STOP_GAINED | LOC_Os07g40510 |
| SBS | Chr7 | 28370149 | C-A | HET | ||
| SBS | Chr8 | 11009887 | C-T | HET | ||
| SBS | Chr8 | 15001720 | G-T | Homo | ||
| SBS | Chr8 | 15214874 | C-T | Homo | ||
| SBS | Chr8 | 21661365 | T-A | HET | ||
| SBS | Chr8 | 21661366 | G-A | HET | ||
| SBS | Chr9 | 10084322 | G-C | HET | ||
| SBS | Chr9 | 3359389 | G-A | HET |
Deletions: 18
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 6510001 | 6537000 | 26999 | 3 |
| Deletion | Chr10 | 6539001 | 6583000 | 43999 | 6 |
| Deletion | Chr10 | 6837001 | 7037000 | 199999 | 24 |
| Deletion | Chr10 | 7092001 | 7158000 | 65999 | 10 |
| Deletion | Chr10 | 7211001 | 7228000 | 16999 | 4 |
| Deletion | Chr10 | 7237001 | 7365000 | 127999 | 20 |
| Deletion | Chr10 | 7370001 | 7620000 | 249999 | 35 |
| Deletion | Chr10 | 7622001 | 7719000 | 96999 | 19 |
| Deletion | Chr11 | 11672078 | 11672096 | 18 | |
| Deletion | Chr8 | 15914891 | 15914892 | 1 | |
| Deletion | Chr8 | 16241757 | 16241770 | 13 | |
| Deletion | Chr2 | 19247096 | 19247107 | 11 | |
| Deletion | Chr2 | 20384518 | 20384524 | 6 | |
| Deletion | Chr8 | 21057699 | 21057705 | 6 | |
| Deletion | Chr5 | 27044699 | 27044702 | 3 | LOC_Os05g46720 |
| Deletion | Chr1 | 27428083 | 27428084 | 1 | |
| Deletion | Chr1 | 39529001 | 39550000 | 20999 | 3 |
| Deletion | Chr1 | 41170923 | 41170924 | 1 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 19077681 | 19077684 | 4 | |
| Insertion | Chr10 | 12235631 | 12235631 | 1 | |
| Insertion | Chr2 | 20911076 | 20911079 | 4 | |
| Insertion | Chr3 | 23143532 | 23143532 | 1 | LOC_Os03g41624 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 13377772 | 13406256 | 2 |
Translocations: 10
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 748146 | Chr11 | 717861 | |
| Translocation | Chr3 | 4463815 | Chr2 | 2674131 | 2 |
| Translocation | Chr12 | 6248354 | Chr8 | 4190745 | |
| Translocation | Chr2 | 7158888 | Chr1 | 35235637 | |
| Translocation | Chr6 | 10800796 | Chr4 | 16769165 | |
| Translocation | Chr10 | 13685211 | Chr6 | 9639506 | 2 |
| Translocation | Chr8 | 17916355 | Chr7 | 22983866 | 2 |
| Translocation | Chr11 | 22304260 | Chr7 | 5845046 | |
| Translocation | Chr11 | 24656029 | Chr8 | 8656763 | 2 |
| Translocation | Chr11 | 25262634 | Chr7 | 5845118 |
