Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2192-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2192-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 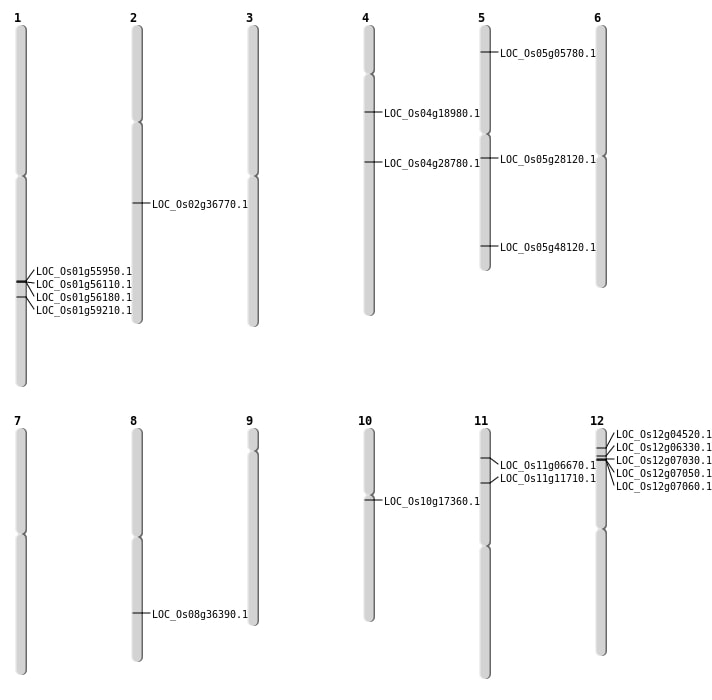
|
| Variant Information |
|---|
Single base substitutions: 59
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 12431493 | C-T | HET | ||
| SBS | Chr1 | 13119810 | T-A | HET | ||
| SBS | Chr1 | 13239050 | A-T | HET | ||
| SBS | Chr1 | 14999276 | C-A | Homo | ||
| SBS | Chr1 | 30609442 | A-T | Homo | ||
| SBS | Chr1 | 32030749 | T-C | HET | ||
| SBS | Chr1 | 32290610 | C-T | HET | ||
| SBS | Chr1 | 32349438 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g56180 |
| SBS | Chr1 | 34205579 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g59210 |
| SBS | Chr1 | 6684462 | C-G | HET | ||
| SBS | Chr10 | 1477806 | G-C | Homo | ||
| SBS | Chr10 | 15375525 | C-A | HET | ||
| SBS | Chr10 | 18663409 | C-T | HET | ||
| SBS | Chr12 | 15795235 | T-C | Homo | ||
| SBS | Chr12 | 1926211 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os12g04520 |
| SBS | Chr12 | 20955399 | T-C | HET | ||
| SBS | Chr12 | 23900576 | G-A | HET | ||
| SBS | Chr12 | 2844445 | C-T | HET | ||
| SBS | Chr2 | 20212545 | G-A | HET | ||
| SBS | Chr2 | 31563083 | G-A | Homo | ||
| SBS | Chr2 | 3252926 | A-T | HET | ||
| SBS | Chr2 | 846481 | T-A | HET | ||
| SBS | Chr2 | 9591495 | A-T | HET | ||
| SBS | Chr3 | 14029113 | G-A | HET | ||
| SBS | Chr3 | 14029114 | G-A | HET | ||
| SBS | Chr3 | 17323063 | C-A | HET | ||
| SBS | Chr3 | 20510641 | T-A | HET | ||
| SBS | Chr3 | 23783393 | T-A | Homo | ||
| SBS | Chr3 | 27138357 | C-T | HET | ||
| SBS | Chr3 | 30108312 | C-T | HET | ||
| SBS | Chr3 | 32460462 | C-T | Homo | ||
| SBS | Chr3 | 32481753 | T-A | Homo | ||
| SBS | Chr4 | 10541708 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os04g18980 |
| SBS | Chr4 | 10848057 | T-A | HET | ||
| SBS | Chr4 | 17045114 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os04g28780 |
| SBS | Chr4 | 22190483 | G-A | HET | ||
| SBS | Chr4 | 27186900 | G-T | HET | ||
| SBS | Chr4 | 30176057 | T-C | Homo | ||
| SBS | Chr4 | 30323560 | T-C | HET | ||
| SBS | Chr4 | 7719942 | G-C | HET | ||
| SBS | Chr5 | 18477212 | G-T | HET | ||
| SBS | Chr5 | 24655954 | G-T | HET | ||
| SBS | Chr5 | 251325 | A-G | HET | ||
| SBS | Chr5 | 28103351 | T-C | HET | ||
| SBS | Chr5 | 29637242 | A-T | Homo | ||
| SBS | Chr6 | 10619881 | A-T | HET | ||
| SBS | Chr6 | 10631448 | G-A | Homo | ||
| SBS | Chr6 | 14390498 | A-G | HET | ||
| SBS | Chr6 | 1631899 | T-C | HET | ||
| SBS | Chr6 | 2517523 | C-A | Homo | ||
| SBS | Chr6 | 326460 | G-A | Homo | ||
| SBS | Chr7 | 13906287 | G-A | HET | ||
| SBS | Chr7 | 29363029 | G-A | HET | ||
| SBS | Chr8 | 18857758 | G-T | HET | ||
| SBS | Chr8 | 7785689 | G-T | HET | ||
| SBS | Chr9 | 12475938 | A-T | HET | ||
| SBS | Chr9 | 1523792 | A-T | HET | ||
| SBS | Chr9 | 17081287 | G-C | HET | ||
| SBS | Chr9 | 6501864 | A-G | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr9 | 1116956 | 1116962 | 6 | |
| Deletion | Chr6 | 2811686 | 2811699 | 13 | |
| Deletion | Chr5 | 2883933 | 2883934 | 1 | LOC_Os05g05780 |
| Deletion | Chr12 | 3438001 | 3465000 | 26999 | 3 |
| Deletion | Chr11 | 4487911 | 4487912 | 1 | |
| Deletion | Chr11 | 5914327 | 5914329 | 2 | |
| Deletion | Chr11 | 6510857 | 6510858 | 1 | LOC_Os11g11710 |
| Deletion | Chr3 | 13499433 | 13499508 | 75 | |
| Deletion | Chr2 | 14986225 | 14986236 | 11 | |
| Deletion | Chr9 | 15153183 | 15153213 | 30 | |
| Deletion | Chr5 | 16452214 | 16452246 | 32 | LOC_Os05g28120 |
| Deletion | Chr3 | 19564324 | 19564479 | 155 | |
| Deletion | Chr2 | 20137537 | 20137541 | 4 | |
| Deletion | Chr1 | 21164388 | 21164389 | 1 | |
| Deletion | Chr2 | 22177793 | 22177800 | 7 | LOC_Os02g36770 |
| Deletion | Chr8 | 22974268 | 22974275 | 7 | LOC_Os08g36390 |
| Deletion | Chr7 | 24507881 | 24507882 | 1 | |
| Deletion | Chr5 | 27588158 | 27588159 | 1 | LOC_Os05g48120 |
| Deletion | Chr3 | 28811875 | 28811881 | 6 | |
| Deletion | Chr3 | 30983940 | 30983945 | 5 | |
| Deletion | Chr3 | 32460426 | 32460432 | 6 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 3190532 | 3190532 | 1 | |
| Insertion | Chr11 | 19527999 | 19528002 | 4 | |
| Insertion | Chr11 | 7448026 | 7448029 | 4 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr12 | 3025584 | 3437628 | 2 |
| Inversion | Chr12 | 3026115 | 3462113 | 2 |
| Inversion | Chr10 | 8748358 | 8761187 | LOC_Os10g17360 |
| Inversion | Chr1 | 32229326 | 32303901 | 2 |
| Inversion | Chr1 | 32229328 | 32303905 | 2 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 3249472 | Chr1 | 2526286 | LOC_Os11g06670 |
| Translocation | Chr12 | 8667522 | Chr11 | 23449782 | |
| Translocation | Chr3 | 27389211 | Chr1 | 32305645 | 2 |
| Translocation | Chr3 | 27389740 | Chr1 | 32305651 | 2 |
| Translocation | Chr11 | 28166031 | Chr6 | 26697238 |
