Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2197-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2197-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 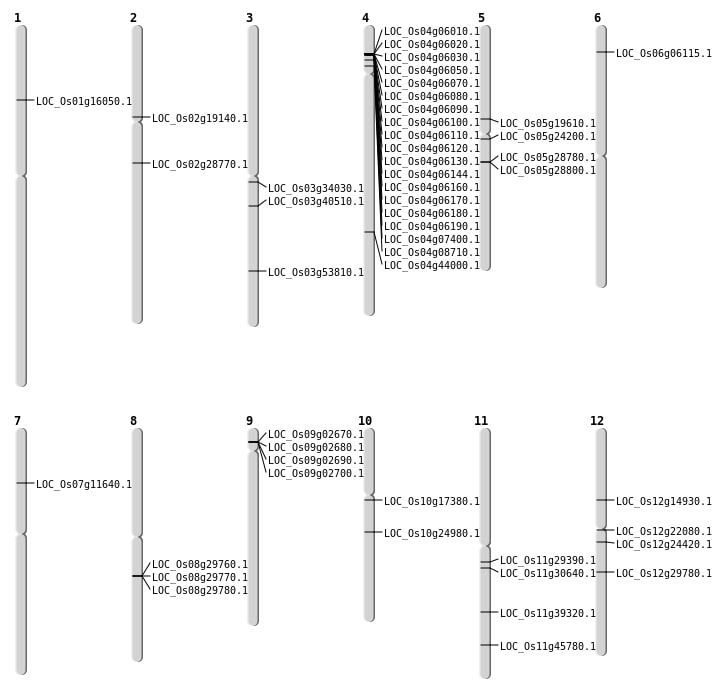
|
| Variant Information |
|---|
Single base substitutions: 30
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 38387671 | A-T | HET | ||
| SBS | Chr10 | 20038810 | G-C | HET | ||
| SBS | Chr10 | 267236 | C-T | HET | ||
| SBS | Chr10 | 2772990 | T-C | HET | ||
| SBS | Chr11 | 11434487 | C-T | HET | ||
| SBS | Chr11 | 17837150 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g30640 |
| SBS | Chr11 | 21168940 | C-T | Homo | ||
| SBS | Chr11 | 23816458 | G-A | Homo | ||
| SBS | Chr12 | 12420907 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22080 |
| SBS | Chr12 | 18201695 | G-C | HET | ||
| SBS | Chr2 | 11158197 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os02g19140 |
| SBS | Chr2 | 2317801 | G-A | Homo | ||
| SBS | Chr2 | 23508454 | C-T | Homo | ||
| SBS | Chr2 | 23785751 | G-C | Homo | ||
| SBS | Chr2 | 2612464 | T-C | Homo | ||
| SBS | Chr2 | 26425164 | A-G | Homo | ||
| SBS | Chr2 | 32974324 | G-A | HET | ||
| SBS | Chr3 | 20722237 | C-T | HET | ||
| SBS | Chr3 | 30866325 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os03g53810 |
| SBS | Chr4 | 11688158 | G-A | Homo | ||
| SBS | Chr4 | 18538213 | A-C | Homo | ||
| SBS | Chr4 | 1867474 | A-T | HET | ||
| SBS | Chr4 | 4728012 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os04g08710 |
| SBS | Chr5 | 14000827 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g24200 |
| SBS | Chr5 | 24204859 | A-G | Homo | ||
| SBS | Chr5 | 26094364 | A-C | HET | ||
| SBS | Chr6 | 25211170 | C-T | HET | ||
| SBS | Chr6 | 7286966 | G-A | HET | ||
| SBS | Chr7 | 22240330 | G-T | HET | ||
| SBS | Chr8 | 16245376 | C-T | HET |
Deletions: 13
Insertions: 12
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 31773232 | 31773233 | 2 | |
| Insertion | Chr1 | 9033670 | 9033670 | 1 | LOC_Os01g16050 |
| Insertion | Chr10 | 10757103 | 10757153 | 51 | |
| Insertion | Chr10 | 164450 | 164450 | 1 | |
| Insertion | Chr11 | 26449357 | 26449357 | 1 | |
| Insertion | Chr2 | 9396604 | 9396604 | 1 | |
| Insertion | Chr4 | 8749036 | 8749036 | 1 | |
| Insertion | Chr4 | 9549950 | 9549950 | 1 | |
| Insertion | Chr5 | 12473144 | 12473144 | 1 | |
| Insertion | Chr7 | 6431611 | 6431616 | 6 | LOC_Os07g11640 |
| Insertion | Chr8 | 2663239 | 2663240 | 2 | |
| Insertion | Chr8 | 3980778 | 3980778 | 1 |
Inversions: 5
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr4 | 3119280 | 3939556 | 2 |
| Inversion | Chr5 | 16876085 | 16886724 | 2 |
| Inversion | Chr5 | 16877203 | 16886725 | 2 |
| Inversion | Chr11 | 17009889 | 17054123 | 2 |
| Inversion | Chr11 | 17009897 | 17054132 | 2 |
Translocations: 15
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 4845481 | Chr1 | 35222208 | |
| Translocation | Chr9 | 7359256 | Chr1 | 18682199 | |
| Translocation | Chr9 | 8289890 | Chr6 | 11646602 | |
| Translocation | Chr12 | 8540937 | Chr1 | 39127625 | LOC_Os12g14930 |
| Translocation | Chr10 | 8760208 | Chr3 | 19409934 | 2 |
| Translocation | Chr5 | 11421900 | Chr1 | 15460374 | LOC_Os05g19610 |
| Translocation | Chr10 | 12860090 | Chr2 | 16818187 | LOC_Os10g24980 |
| Translocation | Chr12 | 13936898 | Chr4 | 16502356 | LOC_Os12g24420 |
| Translocation | Chr12 | 17802716 | Chr7 | 11613953 | LOC_Os12g29780 |
| Translocation | Chr4 | 19009379 | Chr2 | 17022674 | 2 |
| Translocation | Chr11 | 23409493 | Chr5 | 29582833 | LOC_Os11g39320 |
| Translocation | Chr11 | 23409500 | Chr5 | 29582335 | LOC_Os11g39320 |
| Translocation | Chr4 | 26079680 | Chr3 | 22522068 | 2 |
| Translocation | Chr8 | 27374378 | Chr6 | 2829531 | 2 |
| Translocation | Chr11 | 27701516 | Chr4 | 33337464 | LOC_Os11g45780 |
