Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2204-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2204-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 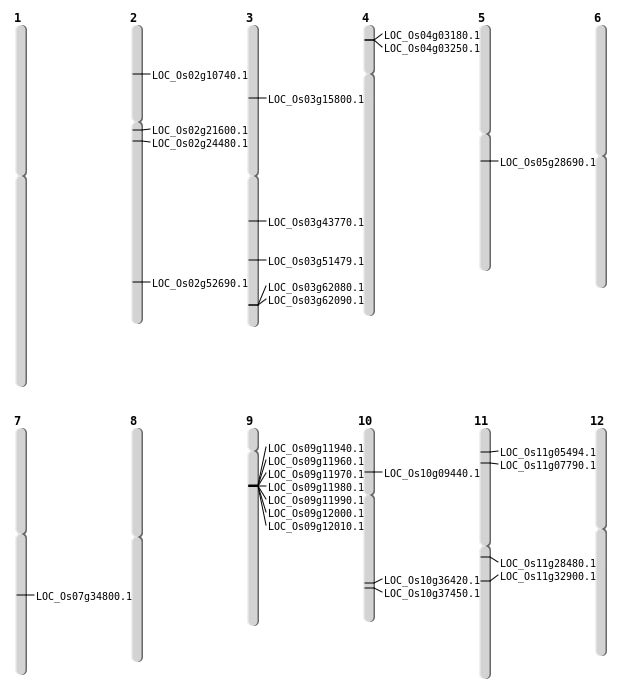
|
| Variant Information |
|---|
Single base substitutions: 57
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 2025928 | G-T | Homo | ||
| SBS | Chr10 | 13594159 | C-A | HET | ||
| SBS | Chr10 | 19471291 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g36420 |
| SBS | Chr10 | 19989949 | A-T | HET | ||
| SBS | Chr10 | 20045642 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os10g37450 |
| SBS | Chr10 | 5092186 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os10g09440 |
| SBS | Chr10 | 6375277 | T-A | HET | ||
| SBS | Chr10 | 6876631 | A-C | HET | ||
| SBS | Chr11 | 11364761 | T-C | HET | ||
| SBS | Chr11 | 11393813 | T-C | HET | ||
| SBS | Chr11 | 13999801 | T-A | HET | ||
| SBS | Chr11 | 15830166 | A-T | HET | ||
| SBS | Chr11 | 161374 | C-T | HET | ||
| SBS | Chr11 | 16791493 | A-C | HET | ||
| SBS | Chr11 | 18342894 | C-G | HET | ||
| SBS | Chr11 | 18680745 | C-T | HET | ||
| SBS | Chr11 | 2481981 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os11g05494 |
| SBS | Chr11 | 28501058 | G-A | HET | ||
| SBS | Chr11 | 3237978 | T-A | HET | ||
| SBS | Chr11 | 3969100 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g07790 |
| SBS | Chr11 | 9438602 | C-A | HET | ||
| SBS | Chr12 | 5533921 | T-C | Homo | ||
| SBS | Chr2 | 12566764 | G-A | HET | ||
| SBS | Chr2 | 32230623 | C-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g52690 |
| SBS | Chr3 | 14176823 | C-T | HET | ||
| SBS | Chr3 | 24489140 | G-C | HET | NON_SYNONYMOUS_CODING | LOC_Os03g43770 |
| SBS | Chr3 | 25464441 | T-C | HET | ||
| SBS | Chr3 | 2766994 | T-G | HET | ||
| SBS | Chr3 | 28118683 | A-G | HET | ||
| SBS | Chr3 | 29479701 | T-A | HET | ||
| SBS | Chr3 | 29479702 | A-T | HET | ||
| SBS | Chr3 | 29479708 | T-G | HET | ||
| SBS | Chr3 | 6097092 | A-G | Homo | ||
| SBS | Chr4 | 1331266 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g03180 |
| SBS | Chr4 | 14533483 | T-C | HET | ||
| SBS | Chr4 | 28993709 | C-T | HET | ||
| SBS | Chr4 | 29470438 | T-G | HET | ||
| SBS | Chr4 | 3272975 | A-G | HET | ||
| SBS | Chr4 | 3346791 | C-T | HET | ||
| SBS | Chr4 | 4523753 | A-T | HET | ||
| SBS | Chr5 | 15305393 | G-A | Homo | ||
| SBS | Chr5 | 22168581 | G-T | HET | ||
| SBS | Chr5 | 6163800 | T-A | HET | ||
| SBS | Chr5 | 6386982 | C-T | HET | ||
| SBS | Chr6 | 10396610 | A-G | Homo | ||
| SBS | Chr6 | 18280784 | A-G | Homo | ||
| SBS | Chr7 | 13671785 | G-C | HET | ||
| SBS | Chr7 | 16880617 | G-A | HET | ||
| SBS | Chr7 | 18863525 | C-A | HET | ||
| SBS | Chr7 | 20856579 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g34800 |
| SBS | Chr7 | 26621083 | A-G | HET | ||
| SBS | Chr9 | 10904392 | C-T | HET | ||
| SBS | Chr9 | 11323508 | G-A | HET | ||
| SBS | Chr9 | 12677576 | G-A | HET | ||
| SBS | Chr9 | 14191705 | G-T | HET | ||
| SBS | Chr9 | 17104685 | C-A | HET | ||
| SBS | Chr9 | 6731309 | C-T | HET |
Deletions: 21
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr1 | 2069714 | 2069718 | 4 | |
| Deletion | Chr6 | 3018811 | 3018824 | 13 | |
| Deletion | Chr2 | 4490106 | 4490112 | 6 | |
| Deletion | Chr2 | 5665742 | 5665747 | 5 | LOC_Os02g10740 |
| Deletion | Chr5 | 5810787 | 5810790 | 3 | |
| Deletion | Chr9 | 6734001 | 6766000 | 31999 | 6 |
| Deletion | Chr3 | 8718468 | 8718478 | 10 | LOC_Os03g15800 |
| Deletion | Chr12 | 8744632 | 8744634 | 2 | |
| Deletion | Chr2 | 12829965 | 12829982 | 17 | LOC_Os02g21600 |
| Deletion | Chr2 | 14179855 | 14179866 | 11 | LOC_Os02g24480 |
| Deletion | Chr12 | 14744978 | 14744979 | 1 | |
| Deletion | Chr7 | 15735148 | 15735156 | 8 | |
| Deletion | Chr11 | 16390369 | 16390376 | 7 | LOC_Os11g28480 |
| Deletion | Chr7 | 18357808 | 18357809 | 1 | |
| Deletion | Chr10 | 19233144 | 19233146 | 2 | |
| Deletion | Chr11 | 25020786 | 25020789 | 3 | |
| Deletion | Chr6 | 27023683 | 27023685 | 2 | |
| Deletion | Chr8 | 27568898 | 27568899 | 1 | |
| Deletion | Chr1 | 30526371 | 30526383 | 12 | |
| Deletion | Chr4 | 31987481 | 31987484 | 3 | |
| Deletion | Chr3 | 35171001 | 35182000 | 10999 | 2 |
Insertions: 3
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr10 | 2066466 | 2066467 | 2 | |
| Insertion | Chr11 | 25044113 | 25044116 | 4 | |
| Insertion | Chr6 | 25246934 | 25246935 | 2 |
Inversions: 6
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr9 | 6721177 | 6731526 | LOC_Os09g11940 |
| Inversion | Chr9 | 6721227 | 6762487 | 2 |
| Inversion | Chr12 | 10524744 | 10650329 | |
| Inversion | Chr12 | 10524745 | 10650334 | |
| Inversion | Chr3 | 28680877 | 28957279 | |
| Inversion | Chr3 | 28957331 | 29455200 | 2 |
Translocations: 4
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr5 | 16830474 | Chr4 | 1383721 | 2 |
| Translocation | Chr5 | 16830497 | Chr4 | 1383622 | 2 |
| Translocation | Chr11 | 19436501 | Chr2 | 526961 | LOC_Os11g32900 |
| Translocation | Chr11 | 19436814 | Chr2 | 526949 | LOC_Os11g32900 |
