Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2207-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2207-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Phenotype |
|
| Mapping (Hover to Zoom-In) | 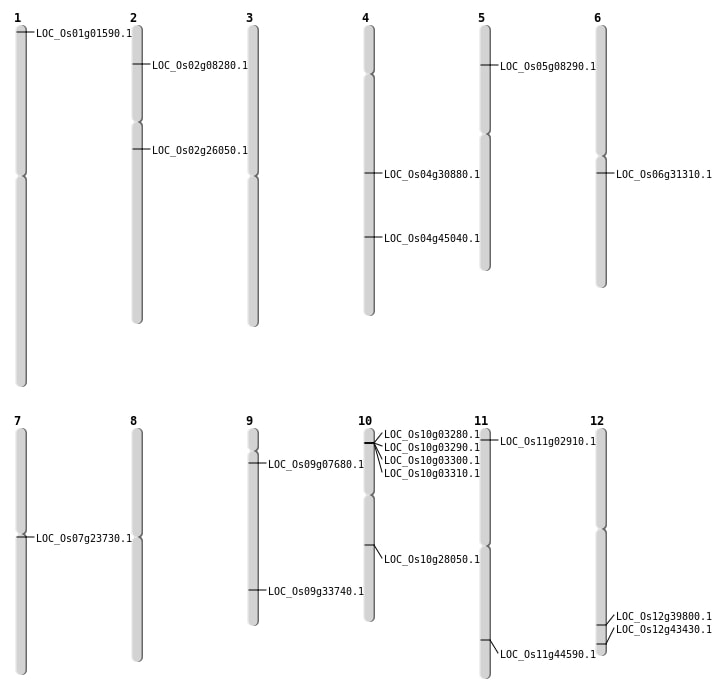
|
| Variant Information |
|---|
Single base substitutions: 51
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 10216982 | T-C | HET | ||
| SBS | Chr1 | 16177106 | A-G | HET | ||
| SBS | Chr1 | 1733394 | G-A | Homo | ||
| SBS | Chr1 | 25147851 | A-T | HET | ||
| SBS | Chr1 | 25247238 | A-T | HET | ||
| SBS | Chr1 | 29344350 | G-C | Homo | ||
| SBS | Chr1 | 3393632 | G-A | Homo | ||
| SBS | Chr1 | 41283106 | G-A | HET | ||
| SBS | Chr1 | 7943915 | G-A | HET | ||
| SBS | Chr10 | 14559604 | T-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os10g28050 |
| SBS | Chr10 | 14749416 | G-C | Homo | ||
| SBS | Chr10 | 6867844 | T-C | HET | ||
| SBS | Chr12 | 10250750 | G-A | Homo | ||
| SBS | Chr12 | 17160627 | C-T | Homo | ||
| SBS | Chr12 | 18221744 | G-A | HET | ||
| SBS | Chr12 | 2236289 | G-C | HET | ||
| SBS | Chr12 | 26256567 | G-A | HET | ||
| SBS | Chr2 | 29665128 | T-G | Homo | ||
| SBS | Chr2 | 31087110 | A-C | HET | ||
| SBS | Chr2 | 32926517 | C-G | Homo | ||
| SBS | Chr2 | 4398262 | G-A | Homo | NON_SYNONYMOUS_CODING | LOC_Os02g08280 |
| SBS | Chr2 | 5322878 | T-A | Homo | ||
| SBS | Chr3 | 1523512 | T-C | HET | ||
| SBS | Chr3 | 1523513 | T-A | HET | ||
| SBS | Chr3 | 15559157 | A-T | HET | ||
| SBS | Chr3 | 34923680 | T-G | HET | ||
| SBS | Chr4 | 16297758 | T-G | HET | ||
| SBS | Chr4 | 18456166 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g30880 |
| SBS | Chr4 | 21152492 | A-G | HET | ||
| SBS | Chr4 | 5442272 | A-T | HET | ||
| SBS | Chr5 | 11304025 | G-A | HET | ||
| SBS | Chr5 | 13618174 | C-T | HET | ||
| SBS | Chr5 | 16163607 | A-T | HET | ||
| SBS | Chr5 | 21062877 | A-G | HET | ||
| SBS | Chr5 | 4527762 | G-T | HET | NON_SYNONYMOUS_CODING | LOC_Os05g08290 |
| SBS | Chr5 | 510701 | T-C | Homo | ||
| SBS | Chr5 | 7939765 | C-T | HET | ||
| SBS | Chr5 | 7939766 | G-T | HET | ||
| SBS | Chr5 | 7939767 | A-G | HET | ||
| SBS | Chr6 | 16428163 | C-T | Homo | ||
| SBS | Chr6 | 28951156 | A-G | Homo | ||
| SBS | Chr6 | 30436203 | G-A | Homo | ||
| SBS | Chr6 | 7675528 | A-G | HET | ||
| SBS | Chr7 | 20783800 | G-A | Homo | ||
| SBS | Chr7 | 7276029 | C-T | HET | ||
| SBS | Chr7 | 7828659 | G-A | Homo | ||
| SBS | Chr7 | 86591 | C-T | HET | ||
| SBS | Chr7 | 86594 | C-T | HET | ||
| SBS | Chr7 | 911784 | A-T | HET | ||
| SBS | Chr7 | 9273667 | T-A | HET | ||
| SBS | Chr8 | 4305909 | G-A | HET |
Deletions: 6
Insertions: 6
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr7 | 19867119 | 20238883 |
Translocations: 13
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr11 | 1001717 | Chr2 | 15292872 | 2 |
| Translocation | Chr7 | 3694488 | Chr4 | 3213549 | |
| Translocation | Chr9 | 3873679 | Chr6 | 18221803 | 2 |
| Translocation | Chr12 | 4227535 | Chr3 | 25611567 | |
| Translocation | Chr7 | 16499425 | Chr6 | 12598602 | |
| Translocation | Chr11 | 19200649 | Chr9 | 4333802 | |
| Translocation | Chr9 | 19929206 | Chr4 | 26648747 | 2 |
| Translocation | Chr11 | 20051526 | Chr6 | 19235822 | |
| Translocation | Chr7 | 22812921 | Chr1 | 293796 | 2 |
| Translocation | Chr12 | 24586754 | Chr1 | 2640511 | LOC_Os12g39800 |
| Translocation | Chr11 | 26252062 | Chr7 | 13389361 | 2 |
| Translocation | Chr12 | 26916650 | Chr2 | 17337440 | LOC_Os12g43430 |
| Translocation | Chr11 | 26960166 | Chr5 | 27709523 | LOC_Os11g44590 |
