Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2260-S [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2260-S Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 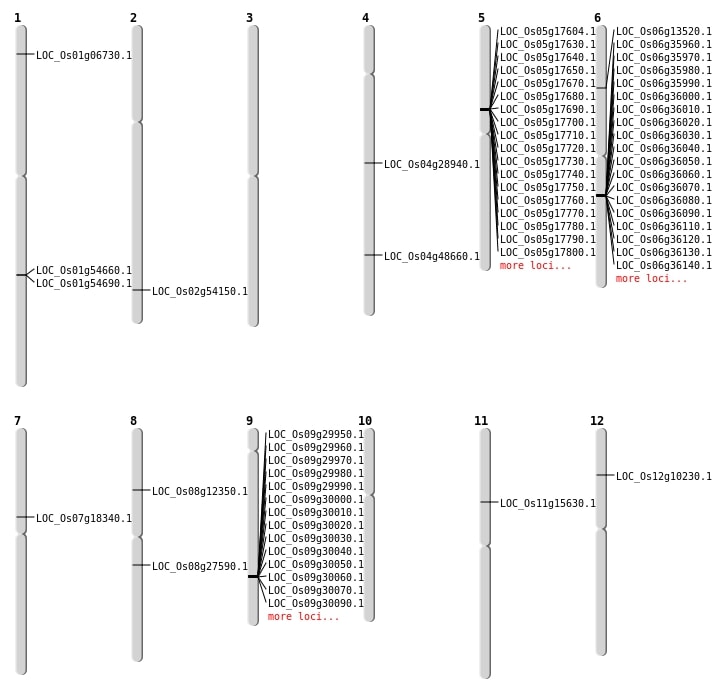
|
| Variant Information |
|---|
Single base substitutions: 58
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 26049036 | T-A | HET | ||
| SBS | Chr1 | 31449024 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54660 |
| SBS | Chr1 | 31467431 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os01g54690 |
| SBS | Chr1 | 36511307 | G-A | HET | ||
| SBS | Chr1 | 9408457 | C-T | Homo | ||
| SBS | Chr10 | 11195667 | A-G | HET | ||
| SBS | Chr10 | 11342721 | C-T | Homo | ||
| SBS | Chr10 | 3900500 | A-G | HET | ||
| SBS | Chr11 | 13724844 | T-C | HET | ||
| SBS | Chr11 | 18629334 | C-T | HET | ||
| SBS | Chr11 | 20176678 | A-T | HET | ||
| SBS | Chr11 | 2398257 | T-A | HET | ||
| SBS | Chr11 | 8386395 | G-T | HET | ||
| SBS | Chr11 | 8422277 | C-A | HET | ||
| SBS | Chr11 | 8869768 | T-A | HET | NON_SYNONYMOUS_CODING | LOC_Os11g15630 |
| SBS | Chr11 | 9071573 | C-G | HET | ||
| SBS | Chr12 | 11405657 | C-T | HET | ||
| SBS | Chr12 | 21690835 | T-A | HET | ||
| SBS | Chr12 | 26277750 | C-T | HET | ||
| SBS | Chr12 | 3947667 | C-G | HET | ||
| SBS | Chr12 | 5401895 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os12g10230 |
| SBS | Chr12 | 7372084 | C-T | HET | ||
| SBS | Chr2 | 11228268 | T-C | HET | ||
| SBS | Chr2 | 27758451 | G-A | HET | ||
| SBS | Chr2 | 5296900 | C-T | HET | ||
| SBS | Chr3 | 12728966 | G-A | HET | ||
| SBS | Chr3 | 16755893 | C-T | Homo | ||
| SBS | Chr3 | 23487007 | G-A | HET | ||
| SBS | Chr3 | 5490724 | A-G | HET | ||
| SBS | Chr3 | 8075410 | C-T | HET | ||
| SBS | Chr4 | 17155976 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os04g28940 |
| SBS | Chr4 | 25215301 | C-G | HET | ||
| SBS | Chr4 | 25891328 | C-T | HET | ||
| SBS | Chr4 | 25891329 | C-T | HET | ||
| SBS | Chr5 | 1357623 | C-T | HET | ||
| SBS | Chr5 | 18856384 | T-A | HET | ||
| SBS | Chr5 | 21655986 | T-C | HET | ||
| SBS | Chr6 | 15196141 | C-A | HET | ||
| SBS | Chr6 | 20156686 | G-T | Homo | ||
| SBS | Chr6 | 23798706 | C-T | HET | ||
| SBS | Chr6 | 25294049 | G-A | HET | ||
| SBS | Chr6 | 7461329 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g13520 |
| SBS | Chr6 | 7563351 | G-A | HET | ||
| SBS | Chr7 | 10868575 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os07g18340 |
| SBS | Chr7 | 21475823 | T-C | HET | ||
| SBS | Chr7 | 22523945 | A-C | HET | ||
| SBS | Chr7 | 227942 | C-T | HET | ||
| SBS | Chr7 | 23287578 | C-T | Homo | ||
| SBS | Chr7 | 2552500 | C-G | HET | ||
| SBS | Chr7 | 9521498 | C-A | Homo | ||
| SBS | Chr8 | 12827599 | G-A | HET | ||
| SBS | Chr8 | 20312105 | C-T | Homo | ||
| SBS | Chr8 | 26313995 | T-C | HET | ||
| SBS | Chr8 | 6696487 | C-A | Homo | ||
| SBS | Chr8 | 6825370 | C-A | HET | ||
| SBS | Chr8 | 6825371 | C-A | HET | ||
| SBS | Chr8 | 7275862 | G-T | Homo | NON_SYNONYMOUS_CODING | LOC_Os08g12350 |
| SBS | Chr9 | 18618317 | C-T | HET |
Deletions: 22
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1686206 | 1686209 | 3 | |
| Deletion | Chr1 | 3184918 | 3184932 | 14 | LOC_Os01g06730 |
| Deletion | Chr6 | 4971883 | 4971899 | 16 | |
| Deletion | Chr7 | 6424672 | 6424687 | 15 | |
| Deletion | Chr6 | 7911870 | 7911878 | 8 | |
| Deletion | Chr5 | 10120001 | 10190000 | 69999 | 9 |
| Deletion | Chr5 | 10193001 | 10266000 | 72999 | 13 |
| Deletion | Chr5 | 10269001 | 10405000 | 135999 | 25 |
| Deletion | Chr6 | 11268941 | 11268943 | 2 | |
| Deletion | Chr6 | 14069412 | 14069415 | 3 | |
| Deletion | Chr8 | 16821797 | 16821801 | 4 | LOC_Os08g27590 |
| Deletion | Chr3 | 17790813 | 17790814 | 1 | |
| Deletion | Chr9 | 18225001 | 18319000 | 93999 | 18 |
| Deletion | Chr1 | 20132441 | 20132444 | 3 | |
| Deletion | Chr11 | 20493593 | 20493599 | 6 | |
| Deletion | Chr6 | 20998001 | 21376000 | 377999 | 41 |
| Deletion | Chr9 | 21145055 | 21145057 | 2 | |
| Deletion | Chr11 | 23268238 | 23268239 | 1 | |
| Deletion | Chr4 | 23764488 | 23764490 | 2 | |
| Deletion | Chr8 | 24619740 | 24619743 | 3 | |
| Deletion | Chr4 | 25869607 | 25869611 | 4 | |
| Deletion | Chr6 | 29708455 | 29708457 | 2 |
Insertions: 5
Inversions: 2
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr2 | 33033902 | 33192681 | 2 |
| Inversion | Chr2 | 33033904 | 33192688 | 2 |
Translocations: 1
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr12 | 18351025 | Chr4 | 29013509 | 2 |
