Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2403 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2403 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 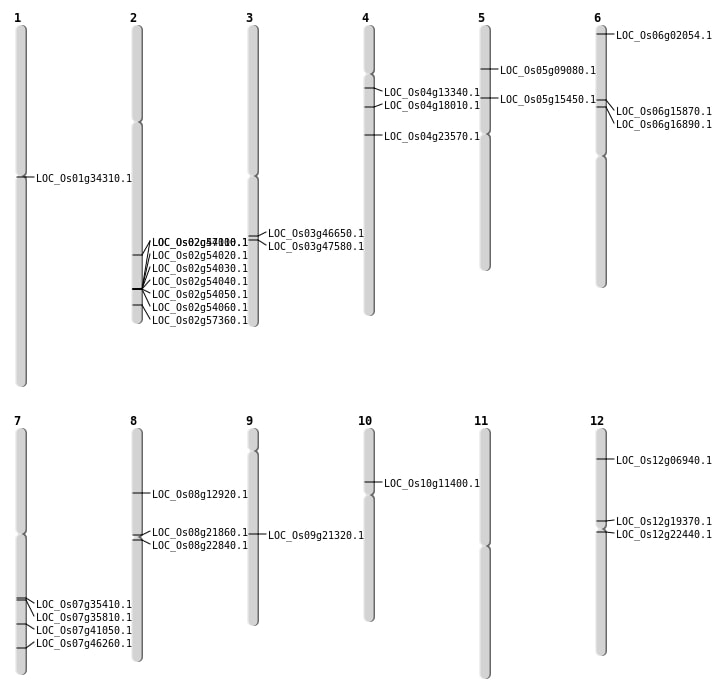
|
| Variant Information |
|---|
Single base substitutions: 79
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 15580472 | A-C | HET | ||
| SBS | Chr1 | 17290453 | T-A | HET | ||
| SBS | Chr1 | 18144992 | G-T | HET | ||
| SBS | Chr1 | 18912901 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os01g34310 |
| SBS | Chr1 | 19287164 | G-A | HET | ||
| SBS | Chr1 | 19287638 | T-C | HET | ||
| SBS | Chr1 | 31743381 | C-T | HET | ||
| SBS | Chr1 | 3770791 | C-T | HET | ||
| SBS | Chr1 | 5051625 | C-A | HET | ||
| SBS | Chr10 | 11483581 | G-A | HET | ||
| SBS | Chr10 | 16159474 | G-T | HET | ||
| SBS | Chr10 | 16864567 | A-T | HET | ||
| SBS | Chr10 | 8361393 | G-T | HET | ||
| SBS | Chr11 | 10044983 | A-T | HET | ||
| SBS | Chr11 | 21924108 | T-C | Homo | ||
| SBS | Chr11 | 7050277 | G-A | HET | ||
| SBS | Chr11 | 9951637 | C-T | HET | ||
| SBS | Chr12 | 11255291 | A-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g19370 |
| SBS | Chr12 | 12430437 | A-T | HET | ||
| SBS | Chr12 | 12433567 | G-A | HET | ||
| SBS | Chr12 | 12671010 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22440 |
| SBS | Chr12 | 15072107 | C-T | HET | ||
| SBS | Chr12 | 15943162 | T-G | HET | ||
| SBS | Chr12 | 26790647 | C-A | HET | ||
| SBS | Chr12 | 3391491 | G-A | HET | STOP_GAINED | LOC_Os12g06940 |
| SBS | Chr12 | 4538793 | C-T | HET | ||
| SBS | Chr12 | 4669108 | C-A | HET | ||
| SBS | Chr2 | 10196783 | T-C | HET | ||
| SBS | Chr2 | 12743571 | G-A | HET | ||
| SBS | Chr2 | 23302969 | C-G | HET | ||
| SBS | Chr2 | 28753567 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os02g47100 |
| SBS | Chr2 | 3516935 | T-C | HET | ||
| SBS | Chr2 | 4644522 | G-T | HET | ||
| SBS | Chr3 | 17796306 | C-T | HET | ||
| SBS | Chr3 | 26824858 | A-G | HET | ||
| SBS | Chr3 | 33473198 | C-T | HET | ||
| SBS | Chr4 | 10057175 | G-A | HET | ||
| SBS | Chr4 | 10857348 | C-A | HET | ||
| SBS | Chr4 | 11115238 | G-A | HET | ||
| SBS | Chr4 | 16266488 | C-T | HET | ||
| SBS | Chr4 | 18925893 | G-T | HET | ||
| SBS | Chr4 | 18973314 | A-T | HET | ||
| SBS | Chr4 | 20648675 | G-T | HET | ||
| SBS | Chr4 | 24807979 | A-C | HET | ||
| SBS | Chr4 | 27874912 | G-A | HET | ||
| SBS | Chr4 | 4053803 | G-C | HET | ||
| SBS | Chr4 | 5865487 | G-T | HET | ||
| SBS | Chr4 | 7416145 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g13340 |
| SBS | Chr5 | 11571564 | C-G | HET | ||
| SBS | Chr5 | 11907260 | C-T | HET | ||
| SBS | Chr5 | 17716771 | C-A | HET | ||
| SBS | Chr5 | 23402724 | C-T | HET | ||
| SBS | Chr5 | 28963008 | G-A | HET | ||
| SBS | Chr5 | 3712833 | C-G | HET | ||
| SBS | Chr5 | 8732598 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os05g15450 |
| SBS | Chr6 | 14088922 | T-A | HET | ||
| SBS | Chr6 | 14088923 | C-A | HET | ||
| SBS | Chr6 | 27430596 | T-A | HET | ||
| SBS | Chr6 | 600827 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os06g02054 |
| SBS | Chr6 | 9013326 | C-A | HET | NON_SYNONYMOUS_CODING | LOC_Os06g15870 |
| SBS | Chr7 | 16443048 | A-G | HET | ||
| SBS | Chr7 | 17369052 | A-T | HET | ||
| SBS | Chr7 | 2078346 | G-T | HET | ||
| SBS | Chr7 | 21340984 | T-C | HET | ||
| SBS | Chr7 | 2360028 | G-A | HET | ||
| SBS | Chr7 | 23617301 | C-T | HET | ||
| SBS | Chr7 | 24556081 | G-A | HET | STOP_GAINED | LOC_Os07g41050 |
| SBS | Chr7 | 29194649 | T-A | HET | ||
| SBS | Chr8 | 13083508 | C-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g21860 |
| SBS | Chr8 | 17700163 | C-A | Homo | ||
| SBS | Chr8 | 20541275 | G-A | HET | ||
| SBS | Chr8 | 6572059 | G-A | HET | ||
| SBS | Chr8 | 7681223 | A-G | HET | NON_SYNONYMOUS_CODING | LOC_Os08g12920 |
| SBS | Chr8 | 899465 | T-A | Homo | ||
| SBS | Chr9 | 11101152 | T-A | HET | ||
| SBS | Chr9 | 12857333 | G-A | HET | STOP_GAINED | LOC_Os09g21320 |
| SBS | Chr9 | 16429170 | A-T | HET | ||
| SBS | Chr9 | 21072838 | C-A | HET | ||
| SBS | Chr9 | 21072839 | G-T | HET |
Deletions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr10 | 6319795 | 6319799 | 4 | LOC_Os10g11400 |
| Deletion | Chr6 | 11818157 | 11818159 | 2 | |
| Deletion | Chr11 | 26698376 | 26698469 | 93 | |
| Deletion | Chr2 | 33079001 | 33118000 | 38999 | 6 |
Insertions: 4
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr1 | 25296082 | 25296083 | 2 | |
| Insertion | Chr12 | 8978787 | 8978799 | 13 | |
| Insertion | Chr3 | 18317751 | 18317752 | 2 | |
| Insertion | Chr7 | 27604320 | 27604322 | 3 | LOC_Os07g46260 |
Inversions: 5
Translocations: 9
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr10 | 6421367 | Chr4 | 4049398 | |
| Translocation | Chr6 | 9792077 | Chr5 | 5048128 | 2 |
| Translocation | Chr4 | 13480180 | Chr2 | 35133159 | 2 |
| Translocation | Chr4 | 13480182 | Chr2 | 35133153 | 2 |
| Translocation | Chr8 | 13733197 | Chr5 | 20095876 | |
| Translocation | Chr8 | 13737081 | Chr5 | 20095885 | LOC_Os08g22840 |
| Translocation | Chr8 | 18930657 | Chr1 | 3645065 | |
| Translocation | Chr8 | 23153088 | Chr6 | 3434471 | |
| Translocation | Chr12 | 27094177 | Chr4 | 9895922 | 2 |
