Search Nipponbare
| Mutant Information |
|---|
| Mutant Name | FN2411 [Download] |
| Generation | M2 |
| Genus Species | Oryza Sativa |
| Cultivar | Kitaake |
| Alignment File (BAM File) | Download FN2411 Alignment File |
| Seed Availability | Yes [Order Seeds] |
| Mapping (Hover to Zoom-In) | 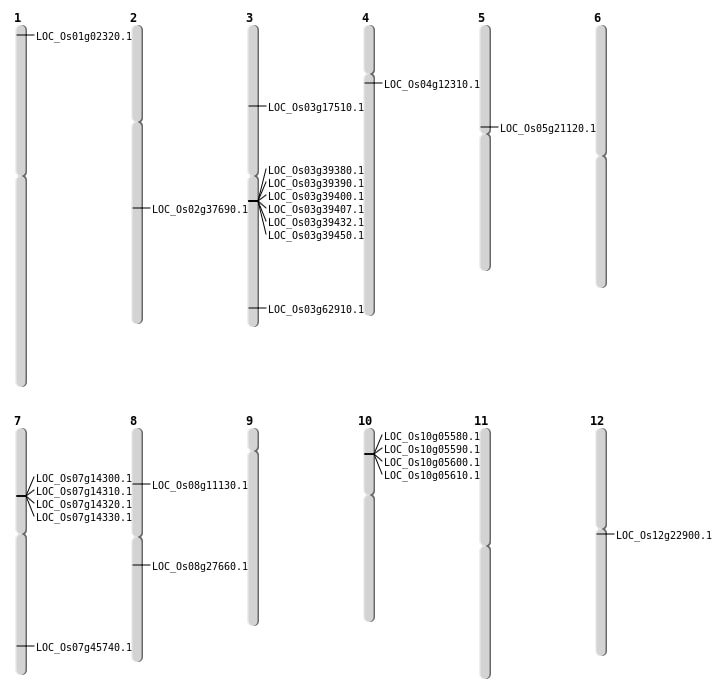
|
| Variant Information |
|---|
Single base substitutions: 65
| Variant Type | Chromosome | Position | Change | Genotype | Effect | Gene Affected |
|---|---|---|---|---|---|---|
| SBS | Chr1 | 14956024 | T-C | HET | ||
| SBS | Chr1 | 25285964 | G-A | HET | ||
| SBS | Chr1 | 32003946 | C-T | HET | ||
| SBS | Chr1 | 34834641 | T-C | HET | ||
| SBS | Chr1 | 36652410 | G-T | HET | ||
| SBS | Chr1 | 5606740 | G-A | HET | ||
| SBS | Chr10 | 11030208 | G-T | HET | ||
| SBS | Chr10 | 13072706 | C-T | HET | ||
| SBS | Chr10 | 4449157 | A-G | Homo | ||
| SBS | Chr10 | 8293284 | G-A | HET | ||
| SBS | Chr11 | 10378174 | G-A | Homo | ||
| SBS | Chr11 | 16044662 | C-G | HET | ||
| SBS | Chr11 | 16696442 | C-T | Homo | ||
| SBS | Chr11 | 27606030 | A-G | HET | ||
| SBS | Chr11 | 3919606 | A-T | HET | ||
| SBS | Chr12 | 12932499 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os12g22900 |
| SBS | Chr12 | 22022165 | G-C | HET | ||
| SBS | Chr12 | 26901771 | C-A | HET | ||
| SBS | Chr12 | 2699832 | C-T | HET | ||
| SBS | Chr12 | 3390656 | T-C | HET | ||
| SBS | Chr12 | 4661693 | G-A | HET | ||
| SBS | Chr12 | 5017858 | T-A | HET | ||
| SBS | Chr2 | 10050107 | C-T | HET | ||
| SBS | Chr2 | 12711457 | C-T | Homo | ||
| SBS | Chr2 | 21554453 | T-A | Homo | ||
| SBS | Chr2 | 2675386 | C-G | HET | ||
| SBS | Chr2 | 26816653 | G-A | HET | ||
| SBS | Chr2 | 6489954 | A-G | HET | ||
| SBS | Chr3 | 2087012 | T-C | HET | ||
| SBS | Chr3 | 31307985 | G-A | HET | ||
| SBS | Chr3 | 36010891 | A-G | HET | ||
| SBS | Chr3 | 9533521 | G-A | HET | ||
| SBS | Chr3 | 9731374 | T-A | HET | STOP_GAINED | LOC_Os03g17510 |
| SBS | Chr4 | 14846897 | C-T | HET | ||
| SBS | Chr4 | 23564109 | A-T | Homo | ||
| SBS | Chr4 | 26873482 | C-T | Homo | ||
| SBS | Chr4 | 33574749 | A-G | Homo | ||
| SBS | Chr4 | 34847283 | G-A | HET | ||
| SBS | Chr4 | 3706242 | C-G | HET | ||
| SBS | Chr4 | 6794737 | C-T | HET | NON_SYNONYMOUS_CODING | LOC_Os04g12310 |
| SBS | Chr4 | 9762905 | T-C | HET | ||
| SBS | Chr5 | 1030528 | C-T | HET | ||
| SBS | Chr5 | 5154448 | C-A | Homo | ||
| SBS | Chr6 | 12619528 | C-T | Homo | ||
| SBS | Chr6 | 17925805 | A-T | Homo | ||
| SBS | Chr6 | 20222800 | C-T | HET | ||
| SBS | Chr6 | 25072192 | A-G | HET | ||
| SBS | Chr6 | 692328 | C-T | HET | ||
| SBS | Chr6 | 8090184 | G-T | HET | ||
| SBS | Chr7 | 14276574 | C-T | HET | ||
| SBS | Chr7 | 2350042 | T-C | Homo | ||
| SBS | Chr7 | 27308020 | G-A | HET | NON_SYNONYMOUS_CODING | LOC_Os07g45740 |
| SBS | Chr8 | 11002016 | G-A | Homo | ||
| SBS | Chr8 | 11937875 | G-A | HET | ||
| SBS | Chr8 | 12304288 | A-T | HET | ||
| SBS | Chr8 | 16351591 | A-T | HET | ||
| SBS | Chr8 | 17462139 | C-T | HET | ||
| SBS | Chr8 | 3891993 | C-T | HET | ||
| SBS | Chr8 | 4718232 | C-T | HET | ||
| SBS | Chr8 | 6546538 | T-C | HET | NON_SYNONYMOUS_CODING | LOC_Os08g11130 |
| SBS | Chr8 | 9601011 | G-A | Homo | ||
| SBS | Chr9 | 11599509 | C-T | HET | ||
| SBS | Chr9 | 15862086 | C-T | HET | ||
| SBS | Chr9 | 17215927 | G-A | HET | ||
| SBS | Chr9 | 7323076 | G-T | Homo |
Deletions: 24
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Deletion | Chr3 | 1088293 | 1088294 | 1 | |
| Deletion | Chr10 | 2771001 | 2802000 | 30999 | 4 |
| Deletion | Chr2 | 4476301 | 4476302 | 1 | |
| Deletion | Chr8 | 4710555 | 4710556 | 1 | |
| Deletion | Chr11 | 7662155 | 7662157 | 2 | |
| Deletion | Chr7 | 8155001 | 8183000 | 27999 | 4 |
| Deletion | Chr4 | 13586372 | 13586390 | 18 | |
| Deletion | Chr6 | 15145253 | 15145274 | 21 | |
| Deletion | Chr9 | 15710872 | 15710877 | 5 | |
| Deletion | Chr9 | 16098466 | 16098467 | 1 | |
| Deletion | Chr8 | 16862583 | 16862589 | 6 | LOC_Os08g27660 |
| Deletion | Chr7 | 17624359 | 17624372 | 13 | |
| Deletion | Chr3 | 20328390 | 20328405 | 15 | |
| Deletion | Chr8 | 21141772 | 21141786 | 14 | |
| Deletion | Chr5 | 21158093 | 21158102 | 9 | |
| Deletion | Chr3 | 21886001 | 21924000 | 37999 | 5 |
| Deletion | Chr12 | 22167302 | 22167319 | 17 | |
| Deletion | Chr2 | 22742957 | 22742971 | 14 | LOC_Os02g37690 |
| Deletion | Chr9 | 22820784 | 22820788 | 4 | |
| Deletion | Chr1 | 23063993 | 23063996 | 3 | |
| Deletion | Chr4 | 25521534 | 25521539 | 5 | |
| Deletion | Chr6 | 30049581 | 30049582 | 1 | |
| Deletion | Chr6 | 30176295 | 30176325 | 30 | |
| Deletion | Chr3 | 33286277 | 33286280 | 3 |
Insertions: 1
| Variant Type | Chromosome | Start | End | Size (bp) | Number of Genes |
|---|---|---|---|---|---|
| Insertion | Chr3 | 35595431 | 35595432 | 2 | LOC_Os03g62910 |
Inversions: 1
| Variant Type | Chromosome | Position 1 | Position 2 | Number of Genes |
|---|---|---|---|---|
| Inversion | Chr1 | 16850876 | 16900505 |
Translocations: 5
| Variant Type | Chromosome | Position 1 | Chromosome | Position 2 | Number of Genes |
|---|---|---|---|---|---|
| Translocation | Chr7 | 8157552 | Chr3 | 21879845 | |
| Translocation | Chr7 | 8182768 | Chr3 | 21882153 | 2 |
| Translocation | Chr5 | 12424813 | Chr1 | 733529 | 2 |
| Translocation | Chr5 | 12427303 | Chr1 | 733526 | 2 |
| Translocation | Chr10 | 17125758 | Chr1 | 41858990 |
